Abstract
Two giant pandas (Ailuropoda melanoleuca) died of unknown causes in a Chinese zoo. The clinical disease profile suggested that the pandas may have suffered a viral infection. Therefore, a series of detection including virus isolation, electron microscopy, cytobiological assay, serum neutralization and RT-PCR were used to identify the virus. It was determined that the isolated virus was a canine coronavirus (CCV), on the basis of coronavirus, neutralization by canine anti-CCV serum, and 84.3% to 100% amino acid sequence similarity with CCV. The results suggest that the affected pandas had been infected with CCV.
The giant panda (Ailuropoda melanoleuca) is an endangered animal that is treasured by humans and strictly protected by law. Currently, pandas face the threat of infectious diseases [3,10]. It has been reported that antibodies against multiple species of viruses such as canine distemper virus, canine coronavirus (CCV) [3,4]. However, little information is available regarding the clinical relevance and epidemiology of these pathogens in giant pandas.
Here we report a strain of CCV isolated from the livers and spleens of diseased pandas.
In July 1997, three adult giant pandas (two 26 year old males and one 12 year old female) in Chongqin city in China, demonstrated roughed hair coats, ataxia, fishy eyed and blurred vision with mucopurulent conjunctivitis followed 5 days later with frothy oral discharge with a fetid odor. 7 days later, the pandas developed an acute onset of dyspnea, vomiting, diarrhea with bloody stools, brown urine and fever. The animals were treated with a combination of cefperazone-sulbactam (80 mg/kg/day for 5 days, IM; Huirui, China) and fluconazole capsules (200 mg per day for 3 weeks, PO; Lanlin, China). After one month of treatment, the 26 year old female and the 12 year old male died with convulsions and other neurologic signs including repetitive muscle fasciculations, muscle stiffening, and collapse. The other female slowly recovered.
Upon necropsy, the lungs, liver, and spleen of all animals had multifocal hemorrhagic foci. Hemorrhage and edema was present in the intestinal mucosa and mesenteric lymph nodes. Sections of liver and spleen were aseptically collected and stored in a 10 mL sterilized glass bottle at -80℃ until used for RNA isolation.
Both of freshly cut surfaces samples of livers and spleens were cultured for bacteriology and incubated in bovine liver medium, ordinary bovine broth, bovine liver agar medium, blood agar medium, and sabouraud glucose agar medium (Beijing Shuangxuan Microbe Culture Medium Products Factory, China), respectively, at 37℃.
The frozen tissues were cut into pieces of about 3 mm and mixed with Hank's solution containing 10% fetal calf serum (FCS; Sigma-Aldrich, USA) at a ratio of 1 : 10 (w/v) and homogenized using a Dounce homogenizer (Beijing Liuyi, China). The homogenates were centrifuged at 3,000 rpm for 30 min, and the supernatant was collected and mixed with 100 units of penicillin (0.01 mL) and 100 units of streptomycin (0.01 mL) per ml before 1 mL of the mixture was covered on the monolayer of Felis catus whole fetus (FCWF) cells, received from the Military Veterinary Institute of Quartermaster University of Chinese People's Liberation Army. The cultures were passed once every 3~4 days until a cytopathic effect (CPE) appeared.
Cells with obvious CPE were collected and diluted with 10 volumes of Hank's solution containing 10% FCS, followed by centrifugation at 3,000 rpm for 10 min to remove large cell debris and at 8,000 rpm for 10 min to remove other particles. The supernatant was subjected to negative-staining as described by Nermut [6] and observed under an electron microscope (TEM, JME-100EA III; JEOL, Japan). Additionally, ultrathin sectioning of cultured cells was used to examine inclusion bodies with reference to a previously described method [8].
The neutralization titre of the isolated virus was determined in FCWF cells and the 50% tissue culture infective dose (TCID50) was calculated using the method of Reed-Muench [1]. The neutralization test was performed as previously described [7]. Briefly, FCWF cells were grown in 96-well flat-bottomed plates until a monolayer of cells formed. Two-fold dilutions of anti-CCV positive or negative serum were added and incubated at 37℃ for 30 min followed by addition with 100 TCID50 of the virus to each well. The neutralization titer is reported as a logarithm of the highest dilution of serum that could neutralize 100 TCID50 of the virus.
Total RNA was extracted from the giant pandas' spleens, infected and non-infected FCWF cells (as positive and negative controls) using the TRizol Reagents kit (Invitrogen, USA) per the manufacturer's recommendations and the RNA samples were stored at -80℃ until use. The extracted total RNA was reverse transcribed to cDNA using avian myeloblastosis virus reverse transcriptase (TaKaRa, Japan) and oligo (dT) primers per the manufacturer's recommendation. The PCR detection was processed according to Naylor et al. [5]. who described a nested PCR methods to identify a CCV. The PCR product was cloned into pGEM-T Easy Vector (Promega, USA) per the manufacturer's recommendations and then sequenced. The sequence was analyzed by GENETYX version 9.0 computer software (Software Development, Japan) and DNAMAN version 4.0 (Lynnon BioSoft, Canada).
Bacteriology failed to grow organisms in cultures inoculated with liver and spleen after 48 h culture.
Cell cultures displayed CPE after 10 passages. Cell fusions could also be observed when comparing infected cultures with negative controls (Figs. 1A and B). The isolated virus was named as giant panda virus (GPV).
To visualize the virus, the culture supernatants were examined under an electron microscope post negative-staining. As shown in Fig. 2A, coronavirus-like viral particles were clearly seen in the supernatant of samples. The ultrathin sections also showed multiple inclusion bodies within the cytoplasm of FCWF cells. Within inclusion bodies some virions had an electron-lucent center, with the nucleocapsid juxtaposed to the envelope, while others were relatively dark when the nucleocapsid was present throughout the particle (Fig. 2B).
A TCID50 of GPV was calculated as 106.30/mL according to Reed-Muench's method [1]. The viral activity of GPV could be neutralized with CCV positive serum from dogs. The mean neutralization titer was 2.18, but when using the CCV negative serum, the neutralization titer was 0.3. This observation demonstrates that the activity of GPV could be specifically neutralized by CCV-positive serum but not by CCV-negative serum.
The virus was further analyzed at the molecular level using RT-PCR. After RT-PCR a 514 bp fragment was amplified from tissues and infected FCWF cells, while PCR for non-infected FCWF cells yielded no results. By sequencing and analysis, the sequences from tissues and infected FCWF cells showed 100% nucleotide identity. As shown in Fig. 3A, the amino acid sequence of the amplified GPV S gene was 98.7% identical to the S protein of CCV K378. In addition, the sequence of the GPV S gene was also 84.3% to 100% identical to the other strains of CCV, including CCV1-71 (AF116246), CCV6 (A22882), CCV C54 (A22886), CCV INSAVC (D13096), UWSMN-1 (AF327928), CCV TN449 (AF116245), and CCV5821 (AB017789) (Fig. 3B).
The pandas in this case study did not respond to anti-bacterial or anti-fungal therapy. However, the clinical course of their disease suggested pathology caused by an infectious organism. Multifocal hemorrhagic foci on tissues including intestine, lung, liver and spleen indicated the organism could induce viremia while severe endosmotic lesions on hepatic lobules and micronodular proliferation of lymphoid cells implied that the organism might propagate in the cells. All of the findings were consistent with a viral disease process. In addition, coronavirus-like viral particles in supernatant from negative-staining spleen and liver tissues were seen under the electron microscope. Therefore, isolation and identification of a potential viral pathogen was pursued.
Canine coronavirus was first isolated from a case of canine enteritis during an epizootic in Germany in 1971. Later, Woods and Wesley [9] reported that CCV could infect neonatal pigs and even older pigs. In addition, Mainka et al. [4] detected antibodies to CCV from captive pandas by neutralization assay. However to date, there have been no published reports of CCV infection in pandas. In this study, we isolated a strain of CCV from two giant pandas, which suggests that pandas can be infected with CCV. To isolate the virus, two inoculation methods including simultaneous inoculation and monolayer-culture inoculation were tried and a coronavirus was successfully isolated. The electron microscopy results provided further evidence that GPV might be a coronavirus-like virus as did the virus neutralization assay.
Next, the GPV sequence was analyzed to further characterize the virus. The results indicated that the amino acid sequence of GPV shared a high identity with other CCV. GPV was most closely related to CCV 1~71 (100% amino acid sequence identity) which was reported as non-fatal to dogs, but may cause a more virulent form of disease in other species [2].
Further characterization of the GPV gene, such as cloning of the entire S gene or other viral structure, may provide additional information on the virus.
Figures and Tables
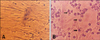 | Fig. 1Isolation and culture of giant panda virus (GPV) in Felis catus whole fetus (FCWF) cell line. (A) Uninfected FCWF cells; (B) FCWF cells inoculated with 10,000 TCID50 GPV. The arrows indicate cells with cytopathic effect, which became round and detatch from the bottom of the culture flask. ×400. |
 | Fig. 2Detection of GPV under electron microscopy. (A) Detection of GPV particles in culture supernatant by negative staining. The arrows indicate the corona spikes of the GPV particles. (B) Detection of inclusion bodies of GPV in FCWF cells by ultrathin section. The white arrow indicates the cytoblast of an infected FCWF cell. The black arrow indicates the inclusion bodies of GPV in the cytoplasm of cells. Scale bars = 200 nm. |
 | Fig. 3Analysis of GPV based on gene sequencing. (A) Comparison of amino acid sequences of S gene product from GPV by nested PCR assay with that of the S gene of canine coronavirus (CCV) K378 (X77047). The asterisk indicates conserved amino acids between GPV and CCV K378; the dot indicates synonymous mutations of amino acids between GPV and CCV K378; the blanks indicate mutant amino acids between GPV to CCV K378; the number after the virus' name corresponds to the amino acid position in the S gene. (B) Percentage of amino acid identity between the partial S gene of GPV and CCV. |
References
1. Glass RT, Bullard JW, Conrad RS, Blewett EL. Evaluation of the sanitization effectiveness of a denture-cleaning product on dentures contaminated with known microbial flora. An in vitro study. Quintessence Int. 2004. 35:194–199.
2. Keenan KP, Jervis HR, Marchwicki RH, Binn LN. Intestinal infection of neonatal dogs with canine coronavirus 1-71: studies by virologic, histologic, histochemical, and immunofluorescent techniques. Am J Vet Res. 1976. 37:247–256.
3. Loeffler IK, Howard J, Montali RJ, Hayek LA, Dubovi E, Zhang Z, Yan Q, Guo W, Wildt DE. Serosurvey of ex situ giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China with implications for species conservation. J Zoo Wildl Med. 2007. 38:559–566.

4. Mainka SA, Qiu X, He T, Appel MJ. Serologic survey of giant pandas (Ailuropoda melanoleuca), and domestic dogs and cats in the Wolong Reserve, China. J Wildl Dis. 1994. 30:86–89.

5. Naylor MJ, Harrison GA, Monckton RP, McOrist S, Lehrbach PR, Deane EM. Identification of canine coronavirus strains from feces by S gene nested PCR and molecular characterization of a new Australian isolate. J Clin Microbiol. 2001. 39:1036–1041.

7. Roche RR, Alvarez M, Guzman MG, Morier L, Kouri G. Comparison of rapid centrifugation assay with conventional tissue culture method for isolation of dengue 2 virus in C6/36-HT cells. J Clin Microbiol. 2000. 38:3508–3510.

8. Tang G, Tang X, Gan D. A method of preparing the section of eye tissue for transmission electron microscopy. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi. 1999. 16:237–239.
9. Woods RD, Wesley RD. Immune response in sows given transmissible gastroenteritis virus or canine coronavirus. Am J Vet Res. 1986. 47:1239–1242.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download