Abstract
The genetics of the prion protein gene (PRNP) play a crucial role in determining the relative susceptibility to transmissible spongiform encephalopathies (TSEs) in several mammalian species. To determine the PRNP gene variability in European red deer (Cervus elaphus), roe deer (Capreolus capreolus) and chamois (Rupicapra rupicapra), the PRNP open reading frame from 715 samples was analysed to reveal a total of ten single nucleotide polymorphisms (SNPs). In red deer, SNPs were found in codons 15, 21, 59, 78, 79, 98, 136, 168 and 226. These polymorphisms give rise to 12 haplotypes, and one of which is identical to the PRNP of American wapiti (Rocky Mountain elk, Cervus elaphus nelsoni). One silent mutation at codon 119 was detected in chamois and no SNPs were found in roe deer. This analysis confirmed that European wild ruminants have a PRNP genetic background that is compatible with TSE susceptibility, including chronic wasting disease.
Chronic wasting disease (CWD) is a transmissible spongiform encephalopathy (TSE) that has occurred in North American cervids for more than 30 years [25]. Natural transmission to humans or domestic livestock seems relatively unlikely, but the possibility still evokes much concern from the public. There is currently no evidence that TSEs (CWD, bovine spongiform encephalopathy, BSE or scrapie) exist in cervids and other wild ruminants in European countries [3,22,24]. Nevertheless, the European Union has implemented active surveillance plans to estimate the prevalence of CWD in the deer population.
An association between variation in the primary sequence of the prion protein gene (PRNP) and disease modulation has been shown for CWD and a number of studies have analysed the genetic variability of the PRNP gene in North American cervid species. Sequence analysis of the PRNP from wapiti (Rocky Mountain elk, Cervus elaphus nelsoni) revealed an amino acid change (Met to Leu) at codon 132: homozygosity for Met132 was then found to be associated with susceptibility [18]. These findings were supported by studies on oral transmission that suggested that the Leu132 may protect against CWD [8]. However, a recent study on CWD cases in free-ranging wapiti showed there was no association of codon 132 with disease susceptibility [20]. Despite this, modulation of the incubation time of CWD by the Leu132 allele is likely [6]. Mule deer (Odocoileus hemionus) that are heterozygous for serine and phenylalanine (S/F heterozygous) or F/F homozygous at codon 225 were underrepresented in the infected population, suggesting there is decreased susceptibility to CWD associated with these genotypes [13]. PRNP polymorphisms encoding amino acid substitutions were identified in white tailed deer (Odocoileus virginianus), with substitutions at residues 65 (G→E), 95 (Q→H), 96 (G→S) and 116 (A→G) [9,19]. Polymorphisms at codons 95, 96 and 116 were found to have a significant influence on the susceptibility to CWD [14,15,19]. Amino acid variations at codons 100 (S→G) and 226 (E→Q) have been described in Chinese and Korean captive sikadeer (Cervus nippon) [11,12]. PRNP genetic variability in European wild ruminants has not been thoroughly investigated, but the identification of PRNP polymorphisms and comparing them with those in North American cervids and domestic small ruminants may provide an estimate of the susceptibility of these species to CWD and other TSEs.
In this study, we describe nucleotide sequence variation in the PRNP locus of red deer (Cervus elaphus), chamois (Rupicapra rupicapra) and roe deer (Capreolus capreolus). A total of 323 red deer samples were collected from Scotland (n = 132; 82 from the mainland and 50 from the Isle of Rhum) and during the hunting seasons from Italy (n = 191). The chamois (n = 203) and roe deer (n = 189) all came from Italy and the sampling covered the whole Alpine arc. Genomic DNA was isolated from blood or frozen muscle tissue with the GenElute Mammalian DNA Kit (Sigma-Aldrich, USA) or the Qiagen Tissue DNA extraction kit (Qiagen, USA). The DNA segments corresponding to the complete open reading frame (ORF) of the PRNP gene (771 bp) were amplified by performing PCR. The primers used for the amplification and sequencing of the deer samples from Scotland were 19fwd (5' ATT TTG CAG ATA AGT CAT C 3'), 778rev (5' AGA AGA TAA TGA AAA CAG GAA G 3') and 315fwd (5' CAG TAA ACC AAA AAC CAA C 3'). A detailed description of the PCR conditions has been published by O'Rourke et al. [18]. The primers used for the wild ruminant samples from Italy were p78 (+) (5' TAA GTG GGC ATA TGA TGC TG 3'), p79 (-) (5' GGG CTG CAG GTA GAC ACT C 3'), p61 (+) (5' AAC CAA CAT GAA GCA TGT GG 3') and p60 (-) (5' GAT AGT AAC GGT CCT CAT AG 3'). PCR was performed according to a previously described protocol [1]. The DNA sequencing reactions were carried out using a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, USA) and the DNA sequences were analysed on an ABI 377 or ABI Prism 3130 Genetic Analyser. Those sequences carrying novel variants have been submitted to the GenBank database under the accession numbers FJ436713-FJ436717. Departures from Hardy-Weinberg equilibrium (HWE) were examined by using the probability test as performed on the GENEPOP software [21]. Unbiased estimates of genotypic disequilibrium were calculated with GENEPOP by using the Markov chain method. The parameters used for all the Markov chain procedures were dememorization of 10,000 steps, 100 batches and 1,000 iterations per batch. The chi-square test for independence was performed in order to establish differences in the allele frequencies between the Scottish and Italian deer. The genetic relationships among the red deer groups at the PRNP locus were estimated using the chord distance (Dc) with the assumption that all the changes of gene frequency were due to genetic drift alone and that the population sizes did not remain constant in all the groups over time [2]. Dendrograms were inferred by the neighbor-joining method [23] with MEGA software [16]. Distance matrices were determined under the assumptions of Kimura's two-parameter model and the confidence values for the individual branches of the resulting tree were determined by bootstrap analysis with 1,000 replicates [4].
In the red deer PRNP, four single-nucleotide polymorphisms (SNPs) encoding amino acid changes were identified: G59S, T98A, P168S and Q226E. Silent mutations were found at codons 15, 21, 78 (which are all novel) (Fig. 1), 79 and 136. The red deer population from the Isle of Rhum was in HWE for all the polymorphisms, while the populations from mainland Scotland and from Italy were not in HWE for SNPs at codons 15 (p < 0.001), 21 (p < 0.001), 226 (p = 0.0014), and 98 (p = 0.03), 136 (p = 0.01), 226 (p = 0.01), respectively. This was not unexpected given that in both cases the sampling was carried out in separated areas and this resulted in structured populations. Tests for genotypic disequilibrium revealed that the same linkage between SNPs at codons 136 and 226 (p < 0.001) was present in all the individual populations.
The haplotypes inferred from the found polymorphisms are reported in Table 1. The detailed allele and genotype frequencies are separately summarized in Tables 2 and 3, according to their geographical origin. The silent mutation at codon 15 (gtg→gcg) was found only in the red deer from mainland Scotland, while the synonymous substitution at codon 21 revealed two allelic variants: gtc→gtt in the red deer from mainland Scotland and gtc→gtg in the Italian red deer. G59S (ggc→agc) and the silent mutation at codon 79 (ccc→cct) were both detected in single animals from Italy. The G59S polymorphism is located in the first PrP peptide repeat by replacing the fourth Gly of the GGGG track. The silent mutation at codon 78 (cag→caa) was found only in Italian red deer. The allele frequencies of the T98A (acc→gcc) polymorphism in all red deer samples were 92.26% and 7.74% for Thr98 and Ala98, respectively. The allele frequencies were calculated by considering the two animal populations separately and according to their geographical origin, and this revealed a significantly different distribution of these alleles (Chi-square = 5.65; p < 0.018). The P168S (cca→tca) polymorphism was found only in one deer from the Isle of Rhum. The combination of the silent mutation at codon 136 (gct→gcc) and the amino acid change Q226E gave origin to all the possible four haplotypes, but c was most frequently on a haplotype with glutamic acid and t was linked to glutamine (Table 1). However, the haplotype distribution in the studied populations appeared to be different with the most frequent haplotype being c136-E226 (65.91%) in the Scottish red deer while the t136-Q226 haplotype prevailed in the Italian animals (59.17%). This observation was confirmed by statistical analysis (Chi-square = 38.27; p < 1 × 10-7). The c136-Q226 and t136-E226 haplotypes were found only in Scottish deer at low frequencies (1.40% and 0.15%, respectively).
The Dc-based analysis of the genetic distances reflected the geographical separation of the three red deer groups included in this study. As expected, the highest genetic distance value was between the red deer from Italy and mainland Scotland (0.089). The distance value between the Italian and Isle of Rhum deer was 0.078 and that between Mainland Scotland and Isle of Rhum deer was 0.059.
In the chamois, sequencing analysis revealed the presence of one synonymous mutation at codon 119 (gct→gcc). This polymorphism was found only in heterozygosis and the allele frequency was 7.39%. No variation was found in the roe deer PRNP and all the sequences were identical to those already deposited in the GenBank database. All the animals of the three species included in the present study possessed five octapeptide repeats. The phylogenetic tree resulting from the alignment of the wild ruminant PRNP sequences, together with the reported sequences of other wild and domestic ungulates, is represented in Fig. 2. The PRNP sequences revealed high similarity among the red deer, roe deer and American cervids and among the sheep, goat and chamois with the result that they were grouped in two separate clusters of the inferred dendrogram. Furthermore, these two groups were more similar to each other than to bovine, which had the most divergent PRNP of the considered species.
Our results confirmed that the red deer PRNP carries several polymorphic sites, and these give rise to different variants of the mature PrP. However, the PRNP polymorphisms associated with an increased susceptibility to CWD in North American cervids were not found in the European deer. All the animals analysed in this study were methionine homozygous at codon 132. The haplotype variant 10 is identical with the PRNP of wapiti, which would make these carriers susceptible to CWD. The mutations at codons 168 and 226 of red deer are located in the β-sheet 2 and the α-helix 3, respectively [26]. Helix 3 has specific properties regarding protein stability, and any amino acid change could imply a different electric charge and a subsequently different conformation of PrPC protein. The Q226E polymorphism is adjacent to codon 225, which is associated with CWD susceptibility in mule deer. In sheep, a proline to leucine substitution at codon 168 has been associated with increased resistance to experimental BSE inoculation [5]. A modulatory role on susceptibility could be hypothesised for polymorphisms at codons 168 and 226 following the exposition of red deer to TSEs. Despite the considerable number of animals analysed in the present study, the PRNP sequence variability was limited to one synonymous polymorphism in chamois and no SNPs were detected in roe deer, showing that the PRNP is conserved in these two species, at least in Italy. This finding is quite unexpected considering the PRNP variability of the other phylogenetically close species. Roe deer belong to Cervidae, the same family as red deer and American cervids whose PRNP coding region has revealed several silent and coding SNPs. Chamois belong to the family Bovidae, subfamily Caprinae, to which sheep and goats also belong, and these two species are characterized by highly polymorphic PRNP coding regions. The sequence alignment of the chamois PRNP with the ovine and caprine PRNPs showed high inter-species homology with a percentage identity of 99.7% and 99.4%, respectively. Even the bovine PRNP coding region, which has low genetic variability, is not as conserved as the chamois PRNP coding region.
The PRNP genetics in European wild ruminants raises interest not only for the possible introduction/circulation of the CWD agent among wild species, but also because of the widespread presence of scrapie across Europe and consequently the possible exposure of wild ruminants to the scrapie prion. There is evidence for occasionally sharing of mountain grazes by European wild ruminants with sheep and goats during the summer season. The TSE agent has the ability to cross the species barrier [10,17] and a hypothesis for the transmission of the scrapie agent to wild ruminants could be reasonably formulated. Indeed, six cases of scrapie were confirmed in two separately maintained flocks of moufflon, and the disease appeared to be endemic in both flocks [27]. The moufflon (Ovis musimon) is one of four types of primitive sheep that are of the same genus as domestic sheep. The analysis of PRNP phylogenesis is commonly used as a criterion for predicting a species' susceptibility to prions and based on our results particular attention should be given to the potential scrapie susceptibility of chamois, which has a PrP amino acid sequence identical to that of sheep and it is clustered with sheep and goat even closer than is moufflon. As expected, red deer and roe deer have a PRNP gene that is closer to the species in which CWD occurs. Additionally, transmission studies have also shown that intracerebral inoculation of a sheep scrapie agent into wapiti resulted in spongiform encephalopathy with accumulations of pathological PrP in the central nervous system [7]. The preliminary data from transgenic mice expressing CerPrPC- L132 suggests that sheep scrapie isolate SSBP/1 is able to overcome the protective effect of the L132 allele [6]. Thus, the association of PRNP polymorphisms with TSE disease could be quite unexpected for wild ruminant species due to the interaction of different hosts to agent-strain combinations.
In conclusion, the genetic analysis carried out in the present study shows that European wild ruminants can be susceptible to TSEs. This highlights the importance of surveillance programmes and it suggests that these species have a possible role as reservoirs for livestock prion diseases.
Figures and Tables
 | Fig. 1Novel noncoding single nucleotide polymorphisms (SNPs) at codons 15, 21 and 78 in the red deer PRNP. Electropherograms show one homozygous (codon 15) and three heterozygous (codons 21 and 78) genotypes. The SNP at codon 15 was detected only in homozygosis in this study. |
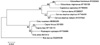 | Fig. 2The phylogenetic tree of similarity among the PRNP gene sequences of the analysed wild ruminants and other wild and domestic ungulates. The following representatives of the suborder Ruminantia have been included in the analysis: mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus), fallow deer (Cervus dama), mouflon (Ovis musimon), goat (Capra hircus), Alpine ibex (Capra ibex), sheep (Ovis aries) and cow (Bos taurus). The GenBank accession numbers of the reference PRNP sequences are indicated. Bootstraps values > 50 (1,000 replicates) are indicated at the internal nodes. The length of each pair of branches represents the distance between sequence pairs. The scale bar represents the percentage of nucleotide differences. |
Acknowledgments
Funding for this project was provided from the Ministry of Health grant IZSPLV 08/03 RC to S.P. and P.L.A., from the Biotechnology and Biological Sciences Research Council, U.K. to M.P., P.St. and W.G.
References
1. Belt PB, Muileman IH, Schreuder BE, Bos-de Ruijter J, Gielkens AL, Smits MA. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol. 1995. 76:509–517.

2. Cavalli-Sforza LL, Edwards AWF. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967. 19:233–257.

3. De Bosschere H, Saegerman C, Neukermans A, Berkvens D, Casaer J, Vanopdenbosch E, Roels S. First chronic wasting disease (CWD) surveillance of roe deer (Capreolus capreolus) in the northern part of Belgium. Vet Q. 2006. 28:55–60.
4. Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981. 17:368–376.

5. Goldmann W, Houston F, Stewart P, Perucchini M, Foster J, Hunter N. Ovine prion protein variant A (136) R (154) L (168) Q (171) increases resistance to experimental challenge with bovine spongiform encephalopathy agent. J Gen Virol. 2006. 87:3741–3745.

6. Green KM, Browning SR, Seward TS, Jewell JE, Ross DL, Green MA, Williams ES, Hoover EA, Telling GC. The elk PRNP codon 132 polymorphism controls cervid and scrapie prion propagation. J Gen Virol. 2008. 89:598–608.

7. Hamir AN, Miller JM, Cutlip RC, Kunkle RA, Jenny AL, Stack MJ, Chaplin MJ, Richt JA. Transmission of sheep scrapie to elk (Cervus elaphus nelsoni) by intracerebral inoculation: final outcome of the experiment. J Vet Diagn Invest. 2004. 16:316–321.

8. Hamir AN, Gidlewski T, Spraker TR, Miller JM, Creekmore L, Crocheck M, Cline T, O'Rourke KI. Preliminary observations of genetic susceptibility of elk (Cervus elaphus nelsoni) to chronic wasting disease by experimental oral inoculation. J Vet Diagn Invest. 2006. 18:110–114.

9. Heaton MP, Leymaster KA, Freking BA, Hawk DA, Smith TP, Keele JW, Snelling WM, Fox JM, Chitko-McKown CG, Laegreid WW. Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle, and deer. Mamm Genome. 2003. 14:765–777.

10. Li L, Coulthart MB, Balachandran A, Chakrabartty A, Cashman NR. Species barriers for chronic wasting disease by in vitro conversion of prion protein. Biochem Biophys Res Commun. 2007. 364:796–800.

11. Meng LP, Zhao DM, Liu HX, Yang JM, Ning ZY, Wu CD, Han CX. Polymorphisms of the prion protein gene (PRNP) in Chinese domestic sika deer (Cervus nippon hortulorum). Anim Genet. 2005. 36:266–267.

12. Jeong HJ, Lee JB, Park SY, Song CS, Kim BS, Rho JR, Yoo MH, Jeong BH, Kim YS, Choi IS. Identification of single-nucleotide polymorphisms of the prion protein gene in sika deer (Cervus nippon laiouanus). J Vet Sci. 2007. 8:299–301.

13. Jewell JE, Conner MM, Wolfe LL, Miller MW, Williams ES. Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J Gen Virol. 2005. 86:2127–2134.
14. Johnson C, Johnson J, Clayton M, McKenzie D, Aiken J. Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of Wisconsin. J Wildl Dis. 2003. 39:576–581.

15. Johnson C, Johnson J, Vanderloo JP, Keane D, Aiken JM, McKenzie D. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol. 2006. 87:2109–2114.

16. Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004. 5:150–163.

17. Moore RA, Vorberg I, Priola SA. Species barriers in prion diseases--brief review. Arch Virol. 2005. 19:Suppl. 187–202.
18. O'Rourke KI, Besser TE, Miller MW, Cline TF, Spraker TR, Jenny AL, Wild MA, Zebarth GL, Williams ES. PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 1999. 80:2765–2769.
19. O'Rourke KI, Spraker TR, Hamburg LK, Besser TE, Brayton KA, Knowles DP. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. 2004. 85:1339–1346.
20. Perucchini M, Griffin K, Miller MW, Goldmann W. PrP genotypes of free-ranging wapiti (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 2008. 89:1324–1328.

21. Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995. 86:248–249.

22. Roels S, Saegerman C, De Bosschere H, Berkvens D, Gregoire F, Hoyoux A, Mousset B, Desmecht D, Vanopdenbosch E, Linden A. First results of chronic wasting disease (CWD) surveillance in the south-eastern part of Belgium. Vet Q. 2005. 27:98–104.

23. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987. 4:406–425.
24. Schettler E, Steinbach F, Eschenbacher-Kaps I, Gerst K, Muessdoerffer F, Risch K, Streich WJ, Frölich K. Surveillance for prion disease in cervids, Germany. Emerg Infect Dis. 2006. 12:319–322.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download