Abstract
Oculocutaneous albinism type 2 (OCA2) is an autosomal recessive disorder that results from mutations in the P gene, and has approximately 70% function of melanin biosynthesis in the melanocytes. While the overwhelming majority of pigmentation studies have focused on European populations, very little is known about the gene and mechanisms affecting skin lightening in Asian population. The main goal of the study was to test the distribution of three polymorphisms located in a pigmentation candidate gene, OCA2, in a sample of individuals of Koreans (N = 250). The genetic markers were selected for polymorphisms that had an allele frequency difference of at least 30% between East Asian populations and European populations. We investigated Minor Allele Frequencies (MAFs) for each of three polymorphisms within OCA2 and reevaluated the difference of the allele frequency along with populations. MAFs of polymorphisms of OCA2 were presented the different frequency in Korean samples (SNP rs1800414 (His615Arg), A allele = 38.8%, rs74653330 (Ala481Thr), A allele = 0.8% and rs7497270 (intronic polymorphism), C allele = 33.4%). While our results had different distributions to European and Caucasians, they showed similar frequencies with East Asian. This study was to reevaluate the distribution of pigmentation candidate gene in Korean samples, further domestic study will aid in developments of the genetic information on worldwide study.
Go to : 
References
1. Scheman AJ, Ray DJ, Witkop CJ Jr., Dahl MV. Hereditary mucoepithelial dysplasia. Case report and review of the literature. J Am Acad Dermatol. 1989; 21:351–7.
2. Montoliu L, Gronskov K, Wei AH, Martinez-Garcia M, Fernandez A, Arveiler B, et al. Increasing the complexity: new genes and new types of albinism. Pigment Cell Melanoma Res. 2014; 27:11–8.

4. Newton JM, Cohen-Barak O, Hagiwara N, Gardner JM, Davisson MT, King RA, et al. Mutations in the human orthologue of the mouse underwhite gene (uw) underlie a new form of oculocutaneous albinism, OCA4. Am J Hum Genet. 2001; 69:981–8.

5. Rinchik EM, Bultman SJ, Horsthemke B, Lee ST, Strunk KM, Spritz RA, et al. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature. 1993; 361:72–6.

6. Rosemblat S, Sviderskaya EV, Easty DJ, Wilson A, Kwon BS, Bennett DC, et al. Melanosomal defects in melanocytes from mice lacking expression of the pink-eyed dilution gene: correction by culture in the presence of excess tyrosine. Exp Cell Res. 1998; 239:344–52.
7. Biossy RE, Vinson RP, Perry V, James WD, Ortonne JP, Nordlund JJ. Dermatologic manifestations of albinism. Medscape. Available from:. http://emedicine.medscape.com/article/1068184-overview#a6.
8. Suzuki T, Miyamura Y, Tomita Y. High frequency of the Ala481Thr mutation of the P gene in the Japanese population. Am J Med Genet A. 2003; 118A:402–3.

10. Kromberg JG, Castle D, Zwane EM, Jenkins T. Albinism and skin cancer in Southern Africa. Clin Genet. 1989; 36:43–52.

11. Parra EJ, Kittles RA, Shriver MD. Implications of correlations between skin color and genetic ancestry for biomedical research. Nat Genet. 2004; 36:S54–60.

12. Edwards M, Bigham A, Tan J, Li S, Gozdzik A, Ross K, et al. Association of the OCA2 polymorphism His615Arg with melanin content in east Asian populations: further evidence of convergent evolution of skin pigmentation. PLoS Genet. 2010; 6:e1000867.

13. Ang KC, Ngu MS, Reid KP, Teh MS, Aida ZS, Koh DX, et al. Skin color variation in Orang Asli tribes of Peninsular Malaysia. PLoS One. 2012; 7:e42752.

14. Ha TW, Han KH, Son DG, Kim SP, Kim DK. Analysis of loss of heterozygosity in Korean patients with keratoacanthoma. J Korean Med Sci. 2005; 20:340–43.

15. Dubey PK, Goyal S, Mishra SK, Yadav AK, Kathiravan P, Arora R, et al. Association analysis of polymorphism in thyroglobulin gene promoter with milk production traits in riverine buffalo (Bubalus bubalis). Meta Gene. 2015; 5:157–61.

16. Donnelly MP, Paschou P, Grigorenko E, Gurwitz D, Barta C, Lu RB, et al. A global view of the OCA2-HERC2 region and pigmentation. Hum Genet. 2012; 131:683–96.

17. Yuasa I, Harihara S, Jin F, Nishimukai H, Fujihara J, Fukumori Y, et al. Distribution of OCA2∗481Thr and OCA2∗ 615Arg, associated with hypopigmentation, in several additional populations. Leg Med (Tokyo). 2011; 13:215–7.
18. Eaton K, Edwards M, Krithika S, Cook G, Norton H, Parra EJ. Association study confirms the role of two OCA2 polymorphisms in normal skin pigmentation variation in East Asian populations. Am J Hum Biol. 2015; 27:520–25.
19. European Bioinformatics institute and the Wellcome Trust Sanger Institute. Ensemble project [Internet]. Hinxton, United Kingdom: [cited 2016 June 1]. Available from: www.ensembl.org,. http://asia.ensembl.org/Homo_sapiens/Variation/Populati. on?db=core;v=rs74653330;vdb= variation.
20. European Bioinformatics institute and the Wellcome Trust Sanger Institute. Ensemble project [Internet]. Hinxton, United Kingdom: [cited 2016 June 1]. Available from: www.ensembl.org,. http://asia.ensembl.org/Homo_sapiens/Variation/Populati. on?db=core;v=rs7497270;vdb= variation.
Go to : 
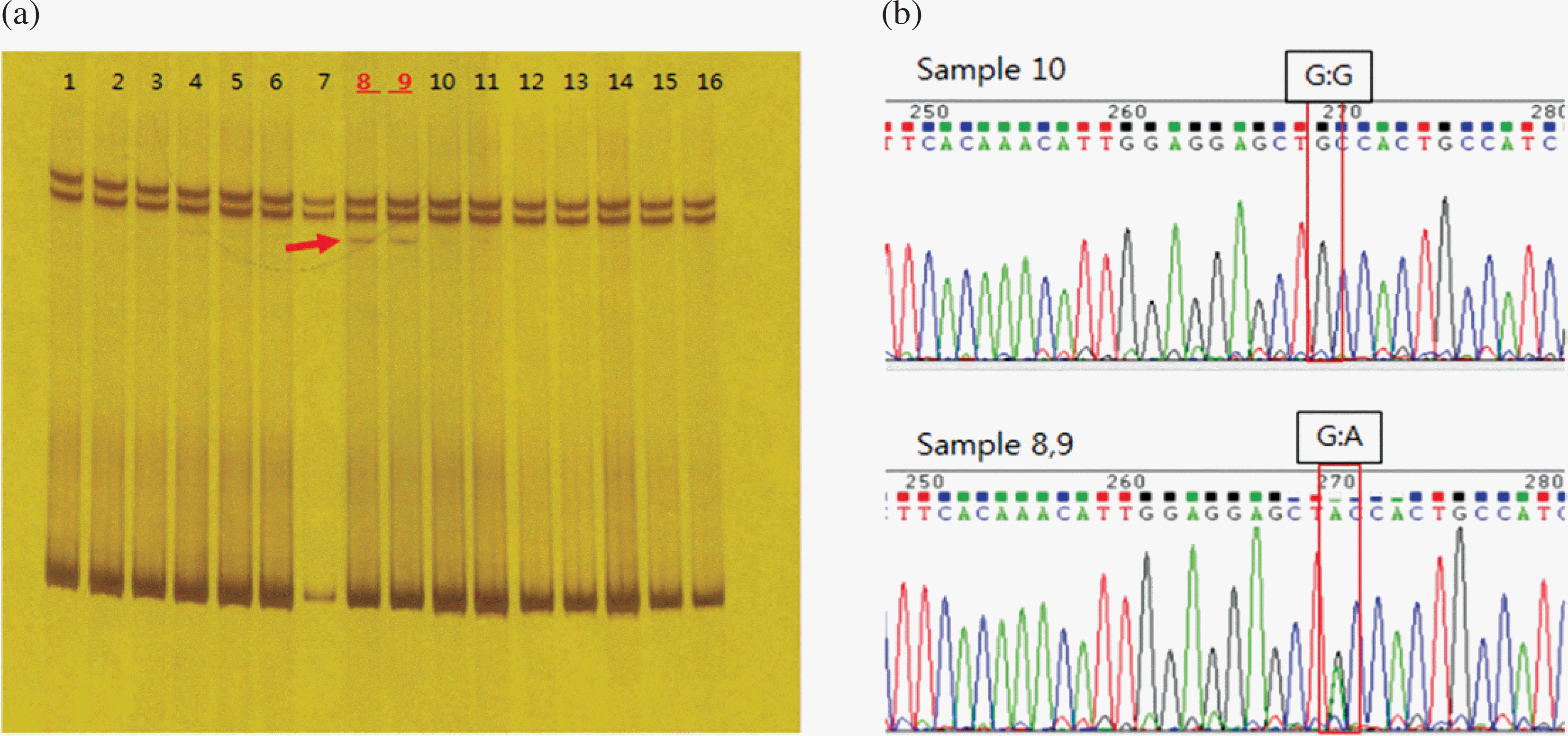 | Fig. 1.Analysis of SSCP and DNA sequences for the SNP rs74653330 (Ala481Thr) within OCA2 in Korean samples. (a) SSCP was performed for the detection of a single-strand DNA fragment by electrophoresis on 10% polyacrylamide gel. (b) DNA sequencing of the SNP (Ala481Thr) region was performed for exon 14 of OCA2 on chromosome 15. |
Table 1.
Primer sequences for PCR-SSCP and detection assay
Table 2.
Genotype frequencies observed for the three polymorphisms analyzed in this study




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download