Abstract
Background
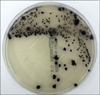
Clostridium difficile is the main etiologic agent of antibiotic-associated diarrhea and the most common cause of hospital-acquired diarrhea. Recently, the incidence of C. difficile infections (CDI) has increased and new highly virulent C. difficile strains have emerged. Therefore, accurate and rapid diagnosis is needed. We compared the results of using chromID C. difficile (chromID CD, bioMeriéux, France) with the conventional C. difficile Selective Agar (CDSA; BD, USA) for the isolation of C. difficile.
Methods
A total of 738 stool specimens of suspected CDI patients at the Severance Hospital from July to August 2011 were inoculated onto CDSA. Among them, 104 stool specimens revealed colonies on CDSA that were then re-inoculated onto chromID CD. The stool samples were stored at -20℃ until the time of the re-inoculation. Cultured agars were interpreted after 24 hrs and 48 hrs, respectively. Species identification was performed on the basis of colony characteristics on agar plates as well as the ATB 32A system (API System SA, France).
References
1. Kyne L, Hamel MB, Polavaram R, Kelly CP. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis. 2002. 34:346–353.
2. Gerding DN, Brazier JS. Optimal methods for identifying Clostridium difficile infections. Clin Infect Dis. 1993. 16:Suppl 4. S439–S442.
3. Fabbro-Peray P, Sotto A, Defez C, Cazaban M, Molinari L, Pinede M, et al. Mortality attributable to nosocomial infection: a cohort of patients with and without nosocomial infection in a French university hospital. Infect Control Hosp Epidemiol. 2007. 28:265–272.
4. Fenner L, Widmer AF, Goy G, Rudin S, Frei R. Rapid and reliable diagnostic algorithm for detection of Clostridium difficile. J Clin Microbiol. 2008. 46:328–330.
5. Shin BM, Kuak EY, Lee EJ, Songer JG. Algorithm combining toxin immunoassay and stool culture for diagnosis of Clostridium difficile infection. J Clin Microbiol. 2009. 47:2952–2956.
6. National Clostridium difficile Standards Group: Report to the Department of Health. J Hosp Infect. 2004. 56:Suppl 1. 1–38.
7. Barbut F, Monot M, Rousseau A, Cavelot S, Simon T, Burghoffer B, et al. Rapid diagnosis of Clostridium difficile infection by multiplex real-time PCR. Eur J Clin Microbiol Infect Dis. 2011. 30:1279–1285.
8. Pai H. Current epidemiology and treatment of Clostridium difficile infection. Infect Chemother. 2010. 42:362–368.
9. Perry JD, Freydiere AM. The application of chromogenic media in clinical microbiology. J Appl Microbiol. 2007. 103:2046–2055.
10. Marler LM, Siders JA, Wolters LC, Pettigrew Y, Skitt BL, Allen SD. Comparison of five cultural procedures for isolation of Clostridium difficile from stools. J Clin Microbiol. 1992. 30:514–516.
11. Kamiya S, Yamakawa K, Ogura H, Nakamura S. Recovery of spores of Clostridium difficile altered by heat or alkali. J Med Microbiol. 1989. 28:217–221.
12. Reller ME, Lema CA, Perl TM, Cai M, Ross TL, Speck KA, et al. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J Clin Microbiol. 2007. 45:3601–3605.
13. Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin Microbiol Infect. 2009. 15:1053–1066.
14. Bloedt K, Riecker M, Poppert S, Wellinghausen N. Evaluation of new selective culture media and a rapid fluorescence in situ hybridization assay for identification of Clostridium difficile from stool samples. J Med Microbiol. 2009. 58:874–877.
15. Delmee M, Van Broeck J, Simon A, Janssens M, Avesani V. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: a plea for culture. J Med Microbiol. 2005. 54:187–191.
16. Lee H, Kim YA, Park KI, Lee K, Chong Y. Detection of toxin B gene of Clostridium difficile by polymerase chain reaction from clinical isolates. Korean J Clin Microbiol. 1999. 2:77–81.
17. Perry JD, Asir K, Halimi D, Orenga S, Dale J, Payne M, et al. Evaluation of a chromogenic culture medium for isolation of Clostridium difficile within 24 hours. J Clin Microbiol. 2010. 48:3852–3858.
18. Nerandzic MM, Donskey CJ. Effective and reduced-cost modified selective medium for isolation of Clostridium difficile. J Clin Microbiol. 2009. 47:397–400.
19. Jang YH, Chung J, Baek S, Park S, Sung H, Kim MN. Implementation of multiplex PCR for species identification and toxin typing in toxigenic Clostridium difficile culture. Korean J Clin Microbiol. 2009. 12:11–16.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download