Abstract
Background
The emergence of non-typhoidal Salmonella (NTS) with decreased susceptibilities to fluoroquinolone, ampicillin, or ceftriaxone has been reported worldwide. However, current surveillance studies of resistance among NTS in Korea are limited. Thus, the antimicrobial susceptibilities; resistance mechanisms such as extended-spectrum β-lactamase (ESBL), plasmid-mediated AmpC β-lactamase (PABL), and plasmid-mediated quinolone resistance (PMQR); and molecular epidemiologic characteristics were investigated in the present study.
Methods
National Institute of Health and National Veterinary Research and Quarantine Service collected NTS strains from 219 clinical and 293 non-clinical specimens from 2006 to 2008. The antimicrobial susceptibilities were determined using the Clinical and Laboratory Standards Institute disk diffusion test. ESBL, PABL, and qnr genotyping were performed using PCR and nucleotide sequencing. Pulsed-field gel electrophoresis was used for the molecular epidemiologic study.
Results
The resistance to ampicillin in clinical and non-clinical NTS was 49% and 18 to 47%, respectively. The resistance rates to trimethoprim-sulfamethoxazole in clinical and non-clinical NTS were 8% and 0 to 41%, respectively. The rates to extended-spectrum cephalosporin were 0 to 1%. One CTX-M-15-producing isolate and four CMY-2-producing isolates were detected. Notably, PFGE analysis showed four isolates carrying blaCMY-2, including one non-clinical strain had high clonality. Although the rate of ciprofloxacin resistance was very low, two qnrS1-carrying NTS strains were detected in non-clinical specimens.
Conclusion
The resistance rates to ampicillin in both clinical and non-clinical NTS were high, while those to trimethoprim-sulfamethoxazole varied depending on the specimen. NTS strains harboring CTX-M-15-type ESBL or CMY-2-type PABL were detected even though the resistance rates to cephalosporins were very low. Four NTS strains carrying the blaCMY-2-gene implied zoonotic infection. Continuous effort to minimize transfer of resistance genes in NTS is necessary.
Figures and Tables
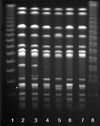 | Fig. 1PFGE band patterns of non-typhoidal Salmonella strains. Lane 1 and 8, size marker; lane 2-3, blaCMY-2-negative non-clinical isolates from swine; lane 4, blaCMY-2-positive non-clinical isolates from swine; lane 5-7, blaCMY-2-positive clinical isolates. All the isolates were S. Typhimurium. |
Table 2
Resistant rates (%) of nontyphoidal Salmonella species to various antimicrobial agents as a result of in vitro antimicrobial susceptibility test

*Clinical isolates included S. Typhimurium (83, 37.9%) and S. Enteritidis (136, 62.1%), †Non-clinical isolates from pig included S. Typhimurium (115, 72.8%), S. Schwarzengrund (18, 11.4%), and S. Rissen (16, 10.1%), ‡Non-clinical isolates from chicken included S. Enteritidis (32, 40.0%), S. Virchow (17, 21.3%), and S. Indiana (9, 11.3%), §Non-clinical isolates from food included S. Enteritidis (22, 40.0%), S. Blokley (19, 34.5%), and S. Typhimurium (9, 13.4%).
Abbreviation: NT, not tested.
References
1. Lee HJ. Salmonellosis. Korean J Clin Microbiol. 2001. 4:5–10.
2. Parry CM, Threlfall EJ. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr Opin Infect Dis. 2008. 21:531–538.
3. Lee K, Yong D, Yum JH, Kim HH, Chong Y. Diversity of TEM-52 extended-spectrum beta-lactamase-producing non-typhoidal Salmonella isolates in Korea. J Antimicrob Chemother. 2003. 52:493–496.
4. Arlet G, Barrett TJ, Butaye P, Cloeckaert A, Mulvey MR, White DG. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 2006. 8:1945–1954.
5. Miriagou V, Tassios PT, Legakis NJ, Tzouvelekis LS. Expanded-spectrum cephalosporin resistance in non-typhoid Salmonella. Int J Antimicrob Agents. 2004. 23:547–555.
6. Centers for Disease Control and Prevention (CDC). Outbreak of multidrug-resistant Salmonella Newport--United States, January-April 2002. MMWR Morb Mortal Wkly Rep. 2002. 51:545–548.
7. Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006. 6:629–640.
8. Wang M, Tran JH, Jacoby GA, Zhang Y, Wang F, Hooper DC. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob Agents Chemother. 2003. 47:2242–2248.
9. Choi SH, Woo JH, Lee JE, Park SJ, Choo EJ, Kwak YG, et al. Increasing incidence of quinolone resistance in human non-typhoid Salmonella enterica isolates in Korea and mechanisms involved in quinolone resistance. J Antimicrob Chemother. 2005. 56:1111–1114.
10. Gay K, Robicsek A, Strahilevitz J, Park CH, Jacoby G, Barrett TJ, et al. Plasmid-mediated quinolone resistance in non-Typhi serotypes of Salmonella enterica. Clin Infect Dis. 2006. 43:297–304.
11. Sjölund-Karlsson M, Folster JP, Pecic G, Joyce K, Medalla F, Rickert R, et al. Emergence of plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates from humans in the United States. Antimicrob Agents Chemother. 2009. 53:2142–2144.
12. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; nineteenth informational supplement. Document M100-S19. 2009. Wayne, PA: CLSI.
13. Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended-spectrum beta-lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect. 2008. 14:Suppl 1. 90–103.
14. Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002. 40:2153–2162.
15. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995. 33:2233–2239.
16. Ran L, Wu S, Gao Y, Zhang X, Feng Z, Wang Z, et al. Laboratory-based surveillance of nontyphoidal Salmonella infections in China. Foodborne Pathog Dis. 2011. 8:921–927.
17. Esaki H, Morioka A, Ishihara K, Kojima A, Shiroki S, Tamura Y, et al. Antimicrobial susceptibility of Salmonella isolated from cattle, swine and poultry (2001-2002): report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. J Antimicrob Chemother. 2004. 53:266–270.
18. Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, et al. Emerging Infections Program NARMS Working Group. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother. 2011. 55:1148–1154.
19. Al-Mashhadani M, Hewson R, Vivancos R, Keenan A, Beeching NJ, Wain J, et al. Foreign travel and decreased ciprofloxacin susceptibility in Salmonella enterica infections. Emerg Infect Dis. 2011. 17:123–125.
20. Yong D, Lim YS, Yum JH, Lee H, Lee K, Kim EC, et al. Nosocomial outbreak of pediatric gastroenteritis caused by CTX-M-14-type extended-spectrum beta-lactamase-producing strains of Salmonella enterica serovar London. J Clin Microbiol. 2005. 43:3519–3521.
21. Tamang MD, Nam HM, Kim TS, Jang GC, Jung SC, Lim SK. Emergence of extended-spectrum beta-lactamase (CTX-M-15 and CTX-M-14)-producing nontyphoid Salmonella with reduced susceptibility to ciprofloxacin among food animals and humans in Korea. J Clin Microbiol. 2011. 49:2671–2675.
22. Song W, Kim JS, Kim HS, Park MJ, Lee KM. Appearance of Salmonella enterica isolates producing plasmid-mediated AmpC beta-lactamase, CMY-2, in South Korea. Diagn Microbiol Infect Dis. 2005. 52:281–284.
23. Sjölund-Karlsson M, Howie R, Rickert R, Krueger A, Tran TT, Zhao S, et al. Plasmid-mediated quinolone resistance among non-Typhi Salmonella enterica isolates, USA. Emerg Infect Dis. 2010. 16:1789–1791.
24. Antunes P, Mourão J, Machado J, Peixe L. First description of qnrS1-IncN plasmid in a ST11 Salmonella Enteritidis clinical isolate from Portugal. Diagn Microbiol Infect Dis. 2011. 69:463–465.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download