Abstract
Background
In the present study, the resistance mechanisms against carbapenems and aminoglycosides for 23 strains of multidrug-resistant Acinetobacter baumannii isolated at a university hospital were investigated.
Methods
The minimal inhibitory concentrations (MICs) were determined via broth microdilution or Etest. The genes encoding OXA-type carbapenemases and 16S rRNA methylase were identified using multiplex PCR, and the amplified products were sequenced. Conjugation experiments were conducted, and an epidemiologic study was performed using enterobacterial repetitive intergenic consensus (ERIC)-PCR.
Results
In the isolates, the MICs of the tested aminoglycosides, including arbekacin, were >1024 μg/ mL; the MICs of aztreonam, cefepime, ceftazidime, and ciprofloxacin ranged from 64 to 128 μg/mL; and the MICs of carbapenem ranged from 32 to 64 μg/ mL, as determined through the broth microdilution test. According to the E-test, the MICs of ampicillin/sulbactam and colistin were 8 and 0.25 to 0.38 μg/ mL, respectively. Sequence analysis confirmed that all of the isolates expressed carbapenemases OXA-23 and OXA-66, as well as armA 16S rRNA methylase. In addition, ISAba1 was identified upstream of the gene encoding OXA-23. OXA-23 and armA were not transferred to Escherichia coli J53 cells in the transconjugation experiments. ERIC-PCR molecular fingerprinting produced a single pattern in all cases.
Go to : 
REFERENCES
1. Villegas MV and Hartstein AI. Acinetobacter outbreaks, 1977∼ 2000. Infect Control Hosp Epidemiol. 2003; 24:284–95.
2. Wilks M, Wilson A, Warwick S, Price E, Kennedy D, Ely A, et al. Control of an outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus colonization and infection in an intensive care unit (ICU) without closing the ICU or placing patients in isolation. Infect Control Hosp Epidemiol. 2006; 27:654–8.
3. Walther-Rasmussen J and H⊘iby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006; 57:373–83.
4. Lee K, Kim MN, Choi TY, Cho SE, Lee S, Whang DH, et al. KONSAR Group. Wide dissemination of OXA-type carbapenemases in clinical Acinetobacter spp. isolates from South Korea. Int J Antimicrob Agents. 2009; 33:520–4.

5. Lee JH, Choi CH, Kang HY, Lee JY, Kim J, Lee YC, et al. Differences in phenotypic and genotypic traits against antimicrobial agents between Acinetobacter baumannii and Acinetobacter genomic species 13TU. J Antimicrob Chemother. 2007; 59:633–9.

6. Zong Z, Lü X, Valenzuela JK, Partridge SR, Iredell J. An outbreak of carbapenem-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in western China. Int J Antimicrob Agents. 2008; 31:50–4.

7. Jeon BC, Jeong SH, Bae IK, Kwon SB, Lee K, Young D, et al. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 beta-lactamase in Korea. J Clin Microbiol. 2005; 43:2241–5.
8. Galimand M, Courvalin P, Lambert T. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob Agents Chemother. 2003; 47:2565–71.
9. Lee H, Yong D, Yum JH, Roh KH, Lee K, Yamane K, et al. Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of Klebsiella pneumoniae and Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis. 2006; 56:305–12.

10. Doi Y, Yokoyama K, Yamane K, Wachino J, Shibata N, Yagi T, et al. Plasmid-mediated 16S rRNA methylase in Serratia marcescens conferring high-level resistance to aminoglycosides. Antimicrob Agents Chemother. 2004; 48:491–6.
11. Bogaerts P, Galimand M, Bauraing C, Deplano A, Vanhoof R, De Mendonca R, et al. Emergence of ArmA and RmtB aminoglycoside resistance 16S rRNA methylases in Belgium. J Antimicrob Chemother. 2007; 59:459–64.

12. Doi Y, Adams JM, Yamane K, Paterson DL. Identification of 16S rRNA methylase-producing Acinetobacter baumannii clinical strains in North America. Antimicrob Agents Chemother. 2007; 51:4209–10.
13. Doi Y, de Oliveira Garcia D, Adams J, Paterson DL. Coproduction of novel 16S rRNA methylase RmtD and metallo-β-lactamase SPM-1 in a panresistant Pseudomonas aeruginosa isolate from Brazil. Antimicrob Agents Chemother. 2007; 51:852–6.
14. Yan JJ, Wu JJ, Ko WC, Tsai SH, Chuang CL, Wu HM, Lu YJ, Li JD. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J Antimicrob Chemother. 2004; 54:1007–12.

15. Marques MB, Brookings ES, Moser SA, Sonke PB, Waites KB. Comparative in vitro antimicrobial susceptibilities of nosocomial isolates of Acinetobacter baumannii and synergistic activities of nine antimicrobial combinations. Antimicrob Agents Chemother. 1997; 41:881–5.

16. La Scola B, Gundi VA, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol. 2006; 44:827–32.
18. Lee K, Chong Y, Shin HB, Kim YA, Yong D, Yum JH. Modified Hodge and EDTA-disk synergy tests to screen metallo-β-lactamase-producing strains of Pseudomonas and Acinetobacter species. Clin Microbiol Infect. 2001; 7:88–91.
19. Shin KS, Son BR, Hong SB, Kim J. Dipicolinic acid-based disk methods for detection of metallo-β-lactamase-producing Pseudomonas spp. and Acinetobacter spp. Diagn Microbiol Infect Dis. 2008; 62:102–5.

20. Yum JH, Yong D, Lee K, Kim HS, Chong Y. A new integron carrying VIM-2 metallo-β-lactamase gene cassette in a Serratia marcescens isolate. Diagn Microbiol Infect Dis. 2002; 42:217–9.

21. Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, et al. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000; 44:891–7.
22. Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, et al. Novel acquired metallo-β-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005; 49:4485–91.
23. Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents. 2006; 27:351–3.

24. Segal H, Garny S, Elisha BG. Is IS(ABA-1) customized for Acinetobacter ? FEMS Microbiol Lett. 2005; 243:425–9.
25. Jacoby GA and Han P. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 1996; 34:908–11.
26. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991; 19:6823–31.
27. Donald HM, Scaife W, Amyes SG, Young HK. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000; 44:196–9.
28. Chu YW, Cheung TK, Chu MY, Lo JY, Dijkshoorn L. OXA-23-type imipenem resistance in Acinetobacter baumannii in Hong Kong. Int J Antimicrob Agents. 2009; 34:285–6.

29. Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis. 2009; 64:185–90.

30. Zhou H, Du XX, Yang Q, Zhou JY, Yu YS, Li LJ. Study on carbapenemase and 16S rRNA methylase of imipenem-resistant Acinetobacter baumannii. Zhonghua Liu Xing Bing Xue Za Zhi. 2009; 30:269–72.
31. Zhou H, Yang Q, Yu YS, Wei ZQ, Li LJ. Clonal spread of imipenem-resistant Acinetobacter baumannii among different cities of China. J Clin Microbiol. 2007; 45:4054–7.
32. Kim JW, Heo ST, Jin JS, Choi CH, Lee YC, Jeong YG, et al. Characterization of Acinetobacter baumannii carrying bla(OXA-23), bla(PER-1) and armA in a Korean hospital. Clin Microbiol Infect. 2008; 14:716–8.
Go to : 
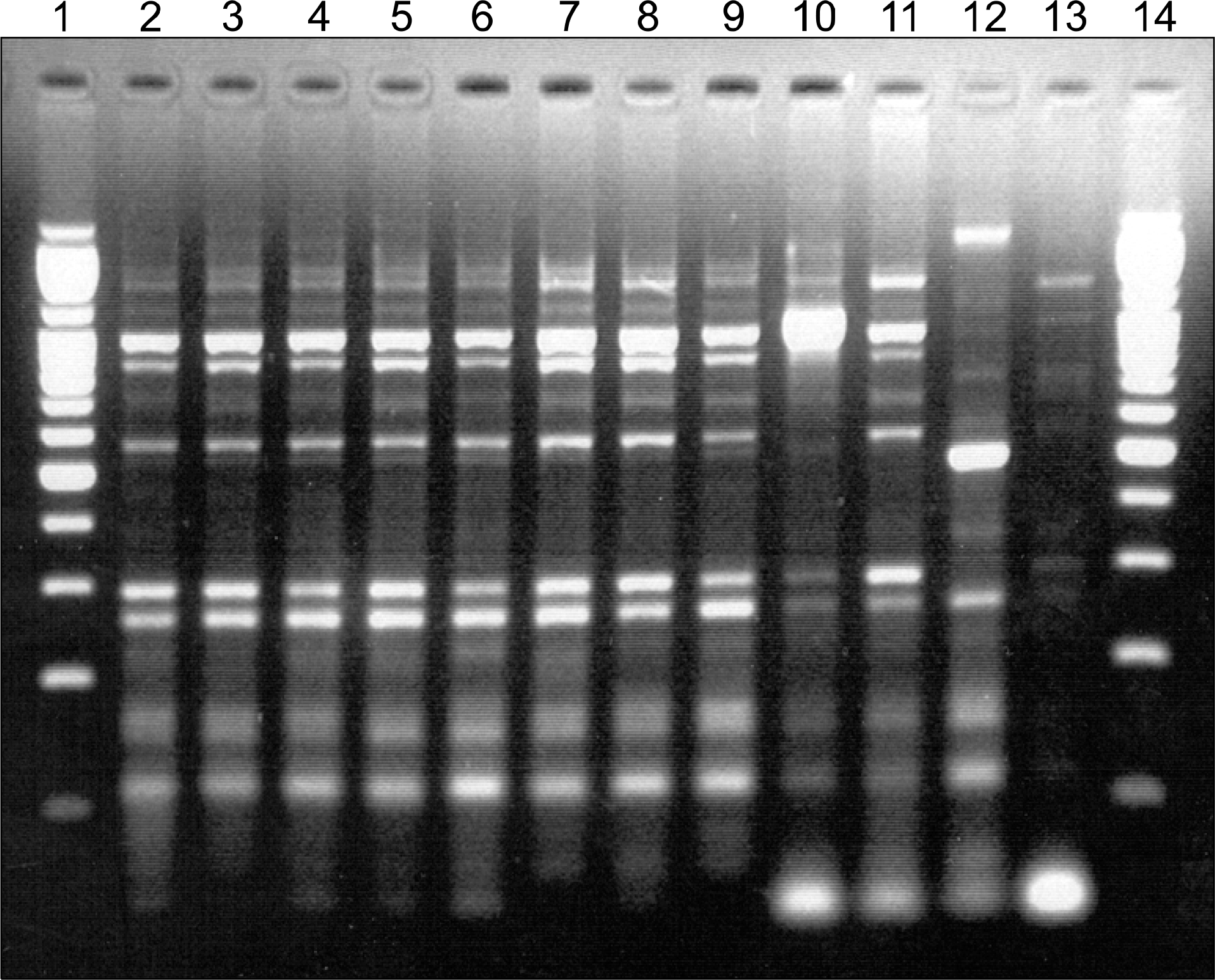 | Fig. 1.The finding of DNA fingerprints by ERIC-PCR of Acinetobacter baumannii clinical isolates. Clinical isolates with outbreak (lanes 2∼9) exhibits single DNA fingerprinting pattern but non-outbreak strains (lanes 10∼13) showed different patterns. Lane 1, molecular size marker (100 bp ladder); lanes 2∼9, clinical isolates of A. baumannii with outbreak; lanes 10∼13, clinical isolates of A. baumannii with non-outbreak strain, including cabapenem resistant A. baumannii isolated from other two University hospital; lane 14, molecular size marker (100 bp ladder). |
Table 1.
Primers used in this study
Table 2.
Minimal inhibitory concentration (MIC) of Acinetobacter baumannii clinical isolates
| No. | Minimal inhibitory concentration (μg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| CTZ | FEP | ATM | A/S∗ | P/T∗ | IMP | MEM | CIP | CS∗ | |
| 88 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 89 | 64 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 91 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 92 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.38 |
| 94 | 128 | 64 | 128 | 8 | >256 | 32 | 32 | 64 | 0.25 |
| 95 | 64 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 96 | 64 | 64 | 128 | 8 | >256 | 32 | 32 | 64 | 0.25 |
| 98 | 64 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 103 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 104 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 105 | 128 | 64 | 128 | 8 | >256 | 64 | 64 | 64 | 0.25 |
| 106 | 64 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 201 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 202 | 128 | 64 | 128 | 8 | >256 | 64 | 64 | 64 | 0.25 |
| 203 | 64 | 64 | 128 | 8 | >256 | 32 | 32 | 64 | 0.25 |
| 205 | 64 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 207 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.38 |
| 208 | 128 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 209 | 128 | 128 | 128 | 8 | >256 | 32 | 32 | 64 | 0.25 |
| 210 | 128 | 128 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 211 | 128 | 128 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 212 | 64 | 64 | 128 | 8 | >256 | 32 | 64 | 64 | 0.25 |
| 213 | 128 | 64 | 128 | 8 | >256 | 32 | 32 | 64 | 0.25 |
Table 3.
Clinical information and genetic characteristics of Acinetobacter baumannii clinical isolates
| No | Isolation date | Isolation ward | Specimen | OXA-like∗ | ISAba1/ OXA-23† | armA‡ | ERIC-PCR | |
|---|---|---|---|---|---|---|---|---|
| 23 | 51 | |||||||
| 88 | 07-05-07 | ICU | Sputum | + | + | + | + | A |
| 89 | 07-05-08 | ICU | Sputum | + | + | + | + | A |
| 91 | 07-05-11 | ICU | Ascitic F | + | + | + | + | A |
| 92 94 | 07-05-15 07-05-28 | ICU ICU | Sputum Ascitic F | + + | + + | + + | + + | AA |
| 95 | 07-05-25 | ICU | Sputum | + | + | + | + | A |
| 96 | 07-05-30 | ICU | Sputum | + | + | + | + | A |
| 98 | 07-06-20 | ICU | Sputum | + | + | + | + | A |
| 103 | 07-07-05 | A-W | Sputum | + | + | + | + | A |
| 104 | 07-07-09 | B-W | Ascitic F | + | + | + | + | A |
| 105 | 07-07-10 | C-W | BA | + | + | + | + | A |
| 106 | 07-07-10 | C-W | BA | + | + | + | + | A |
| 201 | 07-07-19 | ICU | BA | + | + | + | + | A |
| 202 | 07-07-26 | D-W | BA | + | + | + | + | A |
| 203 | 07-07-24 | E-W | Urine | + | + | + | + | A |
| 205 | 07-07-24 | B-W | BA | + | + | + | + | A |
| 207 | 07-09-27 | ICU | Sputum | + | + | + | + | A |
| 208 | 07-09-27 | ICU | Blood | + | + | + | + | A |
| 209 | 07-10-03 | ICU | Sputum | + | + | + | + | A |
| 210 | 07-10-11 | ICU | Sputum | + | + | + | + | A |
| 211 212 | 07-10-11 07-10-13 | ICU E-W | Blood Other | + + | + + | + + | + + | AA |
| 213 | 07-10-29 | ICU | Sputum | + | + | + | + | A |
∗ blaOXA-23 like and blaOXA-51 like was positive by multiplex PCR, and the amplified products were respectively confirmed to OXA-23 and OXA-66 type by sequencing analysis in eight isolates selected arbitrarily.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download