Abstract
Background
Nasal colonization with methicillin-resistant Staphylococcus aureus (MRSA) is a known risk factor for nosocomialtransmission and infection. In an effort to mitigate this problem, topical mupirocin has been widely used for clearing nasal carriage of MRSA. However, mupirocin resistance has become a worldwide concern due to increased use of the antibiotic. The aims of this study were to evaluate the clinical characteristics and prevalence of mupirocin resistance among clinical isolates of staphylococci and to investigate antimicrobial susceptibility.
Methods
A total of 175 S. aureus specimens recovered over a 4-month period from various body sites were tested for resistance to mupirocin and other antibiotics using the Vitek2 automated system. The presence of the mupA gene was assessed in isolates exhibiting resistance to mupirocin and in other selected organisms. The clinical characteristics of the isolates were also reviewed.
Results
Of the 175 S. aureus isolates, 9.1% (16/175) were resistant to mupirocin, with 1.7% (3/175) having high-level resistance (HR) and 7.4% (13/175) having low-level resistance (LR). Patients with HR-mupirocin-resistant S. aureus had a longer duration of hospitalization (P=0.026). Of the 13 LR-mupirocin-resistant S. aureus strains, 11 had identical antibiogram patterns. The mupA gene was detected only among HR isolates.
Go to : 
REFERENCES
1. Muder RR, Brennen C, Wagener MM, Vickers RM, Rihs JD, Hancock GA, et al. Methicillin-resistant staphylococcal colonization and infection in a longterm care facility. Ann Intern Med. 1991; 114:107–12.

2. Klevens RM, Edwards JR, Tenover FC, McDonald LC, Horan T, Gaynes R. National Nosocomial Infections Surveillance System. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin Infect Dis. 2006; 42:389–91.

3. Lee K, Chang CL, Lee NY, Kim HS, Hong SK, Cho HC. Korean nationwide surveillance of antimicrobial. resistance group. Korean nationwide surveillance of antimicrobial resistance of bacteria in 1998. Yonsei Med J. 2000; 41:497–506.
4. Boyce JM. Methicillin-resistant Staphylococcus aureus. Detection, epidemiology, and control measures. Infect Dis Clin North Am. 1989; 3:901–13.
5. Huang SS and Platt R. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin Infect Dis. 2003; 36:281–5.
6. Yang JA, Park DW, Sohn JW, Yang IS, Kim KH, Kim MJ. Molecular analysis of isoleucyl-tRNA synthetase mutations in clinical isolates of methicillin-resistant Staphylococcus aureus with low-level mupirocin resistance. J Korean Med Sci. 2006; 21:827–32.

7. Yanagisawa T, Lee JT, Wu HC, Kawakami M. Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J Biol Chem. 1994; 269:24304–9.

8. Rahman M, Noble WC, Cookson B, Baird D, Coia J. Mupirocin-resistant Staphylococcus aureus. Lancet. 1987; 2(8555):387–8.

9. Bradley SF, Ramsey MA, Morton TM, Kauffman CA. Mupirocin resistance: clinical and molecular epidemiology. Infect Control Hosp Epidemiol. 1995; 16:354–8.

10. Riley TV, Carson CF, Bowman RA, Mulgrave L, Golledge CL, Pearman JW, et al. Mupirocin-resistant methicillin-resistant Staphylococcus aureus in Western Australia. Med J Aust. 1994; 161:397–8.
11. Yun HJ, Lee SW, Yoon GM, Kim SY, Choi S, Lee YS, et al. Prevalence and mechanisms of low- and high-level mupirocin resistance in staphylococci isolated from a Korean hospital. J Antimicrob Chemother. 2003; 51:619–23.

12. Gilbart J, Perry CR, Slocombe B. High-level mupirocin resistance in Staphylococcus aureus: evidence for two distinct isoleucyl-tRNA synthetases. Antimicrob Agents Chemother. 1993; 37:32–8.

13. Pérez-Roth E, López-Aguilar C, Alcoba-Florez J, Méndez-Alvarez S. High-level mupirocin resistance within methicillin-resistant Staphylococcus aureus pandemic lineages. Antimicrob Agents Chemother. 2006; 50:3207–11.
14. Hodgson JE, Curnock SP, Dyke KG, Morris R, Sylvester DR, Gross MS. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob Agents Chemother. 1994; 38:1205–8.

15. Kim EC, Jung HJ, Oh MD, Lee HJ, Oh HS, Choe KW. Epidemiological typing of methicillin-resistant Staphylococcus aureus outbreak isolates by pulsed-field gel electrophoresis and antibiogram. Yonsei Med J. 1998; 39:587–94.
16. Yoo JI, Shin ES, Cha JO, Lee JK, Jung YH, Lee KM, et al. Clonal dissemination and mupA gene polymorphism of mupirocin-resistant Staphylococcus aureus isolates from longterm-care facilities in South Korea. Antimicrob Agents Chemother. 2006; 50:365–7.
17. Boyce JM. Preventing staphylococcal infections by eradicating nasal carriage of Staphylococcus aureus: proceeding with caution. Infect Control Hosp Epidemiol. 1996; 17:775–9.

18. Baird D and Coia J. Mupirocin-resistant. staphylococcus aureus. Lancet. 1987; 2:387–8.
19. Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus: does mupirocin remain effective? Infect Control Hosp Epidemiol. 2003; 24:342–6.
20. Harbarth S, Dharan S, Liassine N, Herrault P, Auckenthaler R, Pittet D. Randomized, placebo-controlled, double-blind trial to evaluate the efficacy of mupirocin for eradicating carriage of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999; 43:1412–6.
21. Wise R and Johnson J. Mupirocin resistance. Lancet. 1991; 338(8766):578.
22. Orrett FA. The emergence of mupirocin resistance among clinical isolates of methicillin-resistant Staphylococcus aureus in Trinidad: a first report. Jpn J Infect Dis. 2008; 61:107–10.
23. Jones JC, Rogers TJ, Brookmeyer P, Dunne WM Jr, Storch GA, Coopersmith CM, et al. Mupirocin resistance in patients colonized with methicillin-resistant Staphylococcus aureus in a surgical intensive care unit. Clin Infect Dis. 2007; 45:541–7.

24. Song W, Lee TJ, Kim SJ, Park MJ, Lee KM. Methicillin-resistant staphylococcus aureus infections in intensive care unit (ICU) patients: relation to nasal carriage of patients or ICU personnels. Korean J Clin Microbiol. 2001; 4:45–51.
25. Ko KS, Park S, Peck KR, Shin EJ, Oh WS, Lee NY, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus spread by neonates transferred from primary obstetrics clinics to a tertiary care hospital in Korea. Infect Control Hosp Epidemiol. 2006; 27:593–7.
26. Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O'Brien FG, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol. 2002; 40:4289–94.
Go to : 
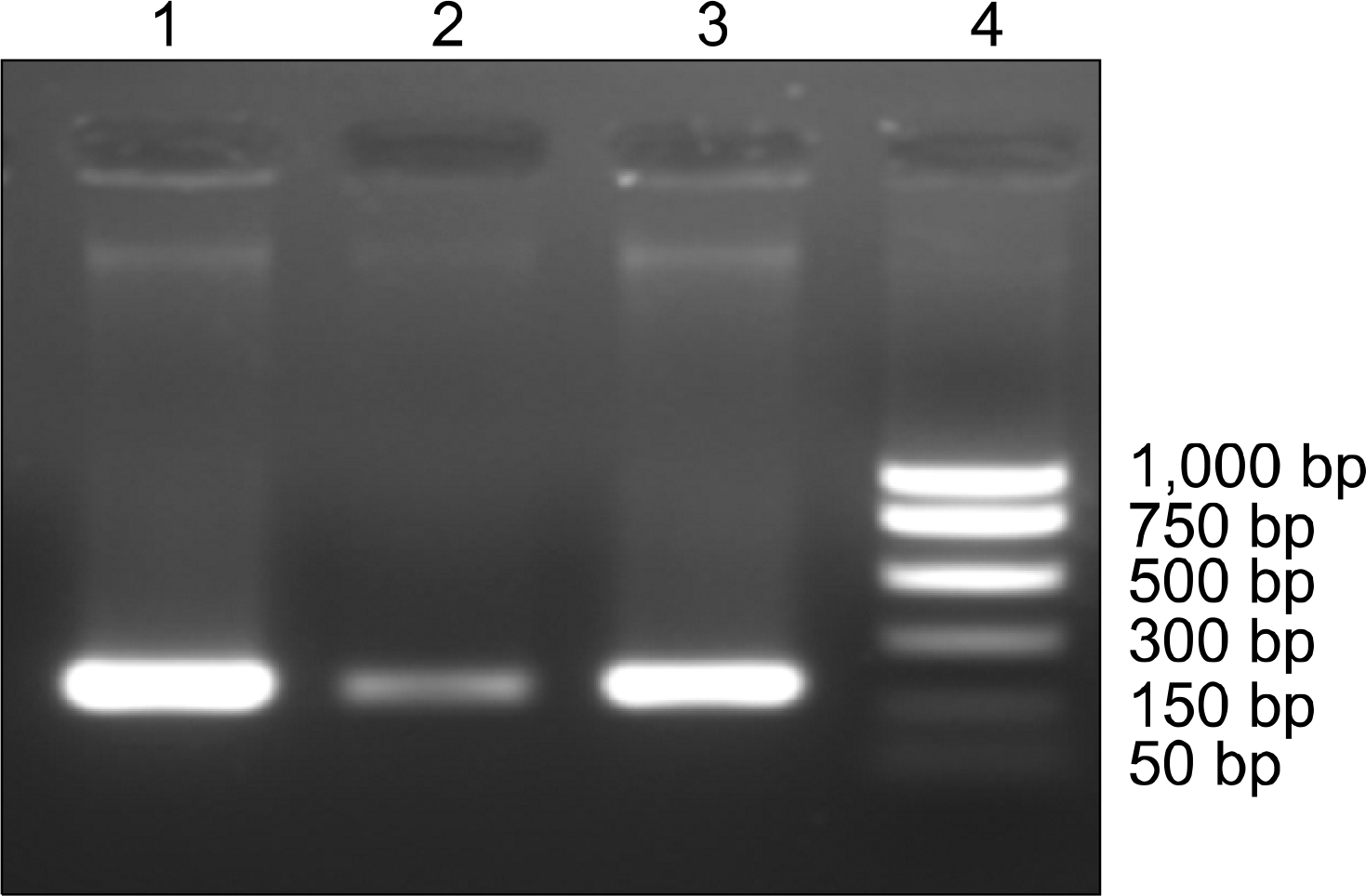 | Fig. 1.Bands of mupA gene in 3 high-level resistance of mupirocin. Lane 1, patient No. 162; Lane 2, patient No. 192; Lane 3, patient No. 195; Lane 4, PCR size marker. |
Table 1.
Distribution of mupirocin MICs among 175 strains of Staphylococcus aureus
| Mupirocin resistance | MIC (μg/mL) | No. of isolate (%) |
|---|---|---|
| Susceptible | <2 | 150 (85.7) |
| 4 | 9 (5.1) | |
| Low-level resistant | 64 | 13 (7.4) |
| High-level resistant | ≥512 | 3 (1.7) |
| Total | 175 (100) |
Table 2.
Clinical characteristics of 175 strains of Staphylococcus aureus
Table 3.
Antimicrobial resistance profile of 108 MRSA strain
Table 4.
Epidemiologic and clinical features of the sixteen mupirocin-resistant Staphylococcus aureus
| No. | MR | Antibiogram∗ | Specimen | Place | Hospita stay (days) | Primary diagnosis |
|---|---|---|---|---|---|---|
| 4 | HR | LQVT | Blood | ICU | 222 | COPD |
| 8 | LR | LQRTsVT | Sputum | GW | 6 | COPD |
| 10 | LR | LQRTsVT | Ascitic fluid | GW | 24 | Duodenal ulcer |
| 15 | LR | GLQRTsVT | Sputum | ICU | 5 | SAH |
| 55 | LR | LQRTsVT | Sputum | ED | 2 | COPD |
| 65 | LR | LQRTsVT | Sputum | GW | 30 | Gangrene of toe |
| 66 | LR | LQRTsVT | Wound | GW | 2 | Sacral sore |
| 79 | LR | LQRTsVT | Sputum | GW | 4 | ESRD |
| 92 | LR | LQVT | Sputum | GW | 9 | Pneumothorax |
| 99 | LR | LQRTsVT | Sputum | GW | 60 | Aplastic anemia |
| 100 | LR | LQRTsVT | Sputum | ICU | 4 | Septic shock |
| 106 | LR | LQRTsVT | Blood | GW | 4 | ESRD |
| 107 | HR | CLQRTsV | T Wound | GW | 24 | Sacral sore |
| 109 | HR | CiCELQRTs TeVT | Sputum | GW | 55 | SAH |
| 153 | LR | LQRTsVT | Wound | GW | 35 | Open wound of ankle and foot |
| 169 | LR | LQRTsVT | Sputum | ICU | 23 | Spine fracture |
∗ Type of antibiogram: LQVT (susceptible to linezolid, quinupristin-dafopristin, vancomycin, teicoplanin), LQRTsVT (susceptible to linezolid, quinupristin-dafopristin, rifampin, trimethoprim-sulfamethoxazole, vancomycin, teicoplanin), GLQRTsVT (susceptible to gentamicin, linezolid, quinupristin-dafopristin, rifampin, trimethoprim-sulfamethoxazole, vancomycin, teicoplanin), CiCELQRTsTeVT (susceptible to ciprofloxacin, clindamycin, erythromycin, linezolid, quinupristin-dafopristin, rifampin, trimethoprim-sulfamethoxazole, tetracycline, vancomycin, teicoplanin).




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download