Abstract
Background
Enteroviruses are the most frequent etiologic agents of aseptic meningitis and are estimated to be the cause of 70% to 90% of viral meningitis cases. Enterovirus diagnosis can be difficult because clinical features vary according to patient immunity and age. The purpose of this study was to evaluate the performance of the realtime nucleic acid sequence-based amplification (NASBA) assay compared to that of the realtime nested RT-PCR assay for enterovirus detection.
Methods
This study was performed on 96 patients suspected of aseptic meningitis based on clinical features. RNA was extracted using NucliSENS EasyMAG and realtime NASBA assay was performed using NucliSENS EasyQ Enterovirus and NucliSENS EasyQ Basic 2. We also executed in-house realtime nested RT-PCR assay for RNA extracted via QIAamp Viral RNA Mini.
Results
The positive rate of realtime NASBA assay was 45.8% for enterovirus detection. The positive rate of first realtime reverse transcription PCR was 22.9% and the second realtime PCR was 57.3%. The concordant rate of the realtime NASBA assay and first realtime reverse transcription PCR was 75.0%. The concordant rate of the realtime NASBA assay and second realtime PCR was 86.5%.
Go to : 
REFERENCES
1. Rotbart HA, Sawyer MH, Fast S, Lewinski C, Murphy N, Keyser EF, et al. Diagnosis of enteroviral meningitis by using PCR with a colorimetric microwell detection assay. J Clin Microbiol. 1994; 32:2590–2.

3. Landry ML, Garner R, Ferguson D. Comparison of the NucliSens Basic kit (Nucleic Acid Sequence-Based Amplification) and the Argene Biosoft Enterovirus Consensus Reverse Transcription-PCR assays for rapid detection of enterovirus RNA in clinical specimens. J Clin Microbiol. 2003; 41:5006–10.

4. Hsiung GD. Picornaviridae. Hsiung GD, Fong CKY and Landry ML, editors. Hsiung's Diagnostic Virology. 4th ed.New Haven: Yale University Press;2006. p. 119–140.
5. Hyypiä T, Auvinen P, Maaronen M. Polymerase chain reaction for human picornaviruses. J Gen Virol. 1989; 70:3261–8.
6. Rotbart HA. Diagnosis of enteroviral meningitis with the polymerase chain reaction. J Pediatr. 1990; 117:85–9.

7. Schlesinger Y, Sawyer MH, Storch GA. Enteroviral meningitis in infancy: potential role for polymerase chain reaction in patient management. Pediatrics. 1994; 94:157–62.

8. Pozo F, Casas I, Tenorio A, Trallero G, Echevarria JM. Evaluation of a commercially available reverse transcription-PCR assay for diagnosis of enteroviral infection in archival and prospectively collected cerebrospinal fluid specimens. J Clin Microbiol. 1998; 36:1741–5.

9. Rotbart HA. Reproducibility of AMPLICOR enterovirus PCR test results. J Clin Microbiol. 1997; 35:3301–2.

10. Nijhuis M, van Maarseveen N, Schuurman R, Verkuijlen S, de Vos M, Hendriksen K, et al. Rapid and sensitive routine detection of all members of the genus enterovirus in different clinical specimens by realtime PCR. J Clin Microbiol. 2002; 40:3666–70.
11. Watkins-Riedel T, Woegerbauer M, Hollemann D, Hufnagl P. Rapid diagnosis of enterovirus infections by realtime PCR on the LightCycler using the TaqMan format. Diagn Microbiol Infect Dis. 2002; 42:99–105.

13. Wang H, Li J, Liu H, Liu Q, Mei Q, Wang Y, et al. Label-free hybridization detection of a single nucleotide mismatch by immobilization of molecular beacons on an agarose film. Nucleic Acids Res. 2002; 30:e61.

14. Heo SR, Jin SK, Chang HE, Park KU, Song JH, Kim EC. Detection of enterovirus in cerebrospinal fluid by realtime nested reverse transcription polymerase chain reaction. Korean J Lab Med. 2006; 26:9–13.

15. Peigue-Lafeuille H, Croquez N, Laurichesse H, Clavelou P, Auma-îitre O, Schmidt J, et al. Enterovirus meningitis in adults in 1999-2000 and evaluation of clinical management. J Med Virol. 2002; 67:47–53.

16. Landry ML, Garner R, Ferguson D. Rapid enterovirus RNA detection in clinical specimens by using nucleic acid sequence-based amplification. J Clin Microbiol. 2003; 41:346–50.

17. Ririe KM, Rasmussen RP, Wittwer CT. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997; 245:154–60.

Go to : 
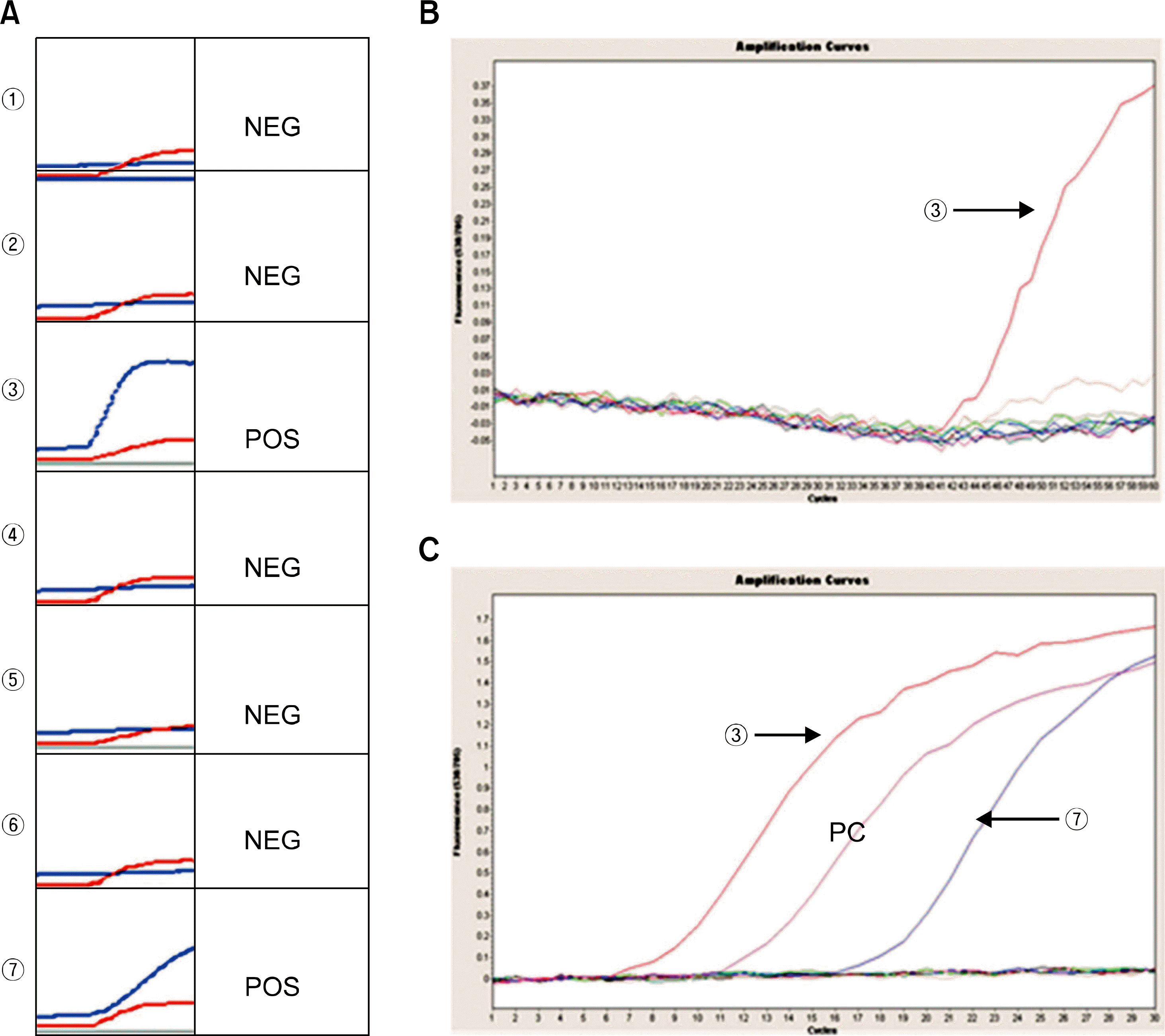 | Fig. 1.Detection of enterovirus by nucleic acid sequence-based amplification and realtime nested reverse transcription PCR. (A) Nucleic acid sequence-based amplification. ①∼ ② and ④∼⑥, samples with a internal control signal (red) and no enterovirus signal (blue); ③ and ⑦, samples with a internal control signal and a strong enterovirus signal. (B) The first realtime reverse transcription PCR. ③, positive for enterovirus. (C) The nested realtime PCR. ③ and ⑦, positive for enterovirus; PC, positive control. |
Table 1.
Comparison of results of enterovirus detection by realtime nucleic acid sequence-based amplification between NucliSENS EasyQ Basic 1 and NucliSENS EasyQ Basic 2
| NucliSENS EasyQ Basic 2 | NucliSENS EasyQ Basic 1 | Total | ||
|---|---|---|---|---|
| Positive | Negative | Invalid | ||
| Positive | 5 | 2 | 7 | |
| Negative | 67 | 8 | 75 | |
| Invalid | 1 | 1 | 2 | |
| Total | 5 | 70 | 9 | 84 |
Table 2.
Comparison of results between realtime nested reverse transcription PCR and realtime nucleic acid sequence-based amplification for detection of enterovirus
| NASBA | First realtime RT-PCR | Nested realtime PCR | Total | ||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Positive | 21 | 23 | 43 | 1 | 44 |
| Negative | 1 | 51 | 12 | 40 | 52 |
| Total | 22 | 74 | 55 | 41 | 96 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download