Abstract
Background
The genes of metallo-β-lactamase (MBL), a powerful carbapenemase, are carried as a part of the mobile gene cassettes inserted into integrons playing an important role in rapid dissemination of antibiotic resistance genes among bacterial isolates. In this study, we investigated carbapenemase genes and class 1 integrons integrated into the gene cassettes in imipenem-non susceptible P. aeruginosa.
Methods
From July 2006 to March 2008, 81 consecutive, non-duplicate, imipenem-non susceptible P. aeruginosa were isolated at Chungnam National University Hospital in Chungcheong province of Korea. The modified Hodge and double disk synergy tests were conducted for the screening of carbapenemase and MBL production, respectively, and PCR and DNA sequencing were performed for the detection of carbapenemase genes and class 1 integron gene cassettes. We also employed the repetitive element sequence-based (Rep)-PCR method for an epidemiologic study.
Results
MBLs were detected in 13.6% (11/81) of imipenem-non susceptible P. aeruginosa. Ten isolates were found to carry blaIMP-1, whereas 1 isolate was found to carry a blaVIM-2. All of the IMP-1-producing strains harbored 4.0 kb class 1 integron containing chloramphenicol, aminoglycoside, and β-lactam-resistant genes. However, blaIMP-1 was not detected at class 1 integron. A 2.5 kb class 1 integron harboring blaVIM-2 was detected in a VIIM-2- producing strain. One identical pattern was observed in ten IMP-1 producing strains.
Go to : 
REFERENCES
1. Bergogne-Berezin E. Pseudomonas and miscellaneous gram-negative bacilli. Cohen J, Powerly WG, editors. eds.Infectious Diseases. New York: Mosby;2004. p. 2203–26.
2. Jacoby GA, Medeiros AA. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991; 35:1697–704.
3. Song W, Woo HJ, Kim JS, Lee KM. In vitro activity of β-lactams in combination with other antimicrobial agents against resistant strains of Pseudomonas aeruginosa. Int J Antimicrob Agents. 2003; 21:8–12.
4. Fernández-Cuenca F, Martinez-Martinez L, Conejo MC, Ayala JA, Perea EJ, Pascual A. Relationship between β-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J Antimicrob Chemother. 2003; 51:565–74.
5. Clark RB. Imipenem resistance among Acinetobacter baumannii: association with reduced expression of a 33-36 kDa outer membrane protein. J Antimicrob Chemother. 1996; 38:245–51.
6. Yu YS, Yang Q, Xu XW, Kong HS, Xu GY, Zhong BY. Typing and characterization of carbapenem-resistant Acinetobacter calcoaceticus-baumannii complex in a Chinese hospital. J Med Microbiol. 2004; 53:653–6.
7. Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001; 45:2598–603.
8. Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991; 35:147–51.
9. Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, et al. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob Agents Chemother. 2002; 46:1053–8.
10. Toleman MA, Simm AM, Murphy TA, Gales AC, Biedenbach DJ, Jones RN, et al. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J Antimicrob Chemother. 2002; 50:673–9.
11. Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob Agents Chemother. 2004; 48:4654–61.
12. Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, et al. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005; 49:4485–91.
13. Bou G, Oliver A, Martinez-Beltrán J. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother. 2000; 44:1556–61.
14. Yoon WS, Lee BY, Bae IK, Kwon SB, Jeong SH, Jeong TJ, et al. Prevalence of imipenem-resistant Pseudomonas aeruginosa isolates and mechanisms of resistance. Korean J Clin Microbiol. 2005; 8:26–33.
15. Kim IS, Oh WI, Song JH, Lee NY. Screening and identification of metallo-β-lactamase gene in clinical isolates of imipenem-resistant Pseudomonas aeruginosa. Korean J Lab Med. 2004; 24:177–82.
16. Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, et al. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000; 44:891–7.
17. Recchia GD, Hall RM. Gene cassettes: a new class of mobile element. Microbiology. 1995; 141:3015–27.

18. Richet HM, Mohammed J, McDonald LC, Jarvis WR. Building communication networks: international network for the study and prevention of emerging antimicrobial resistance. Emerg Infect Dis. 2001; 7:319–22.
19. Ploy MC, Lambert T, Couty JP, Denis F. Integrons: an antibiotic resistance gene capture and expression system. Clin Chem Lab Med. 2000; 38:483–7.

20. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. M100-S10 (M2). Wayne, Pensylvania: CLSI,. 2006.
21. Lee K, Lim YS, Yong D, Yum JH, Chong Y. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2003; 41:4623–9.
22. Kim JM, Kang HK, Jeong SH, Bae IK, Kwon SB, Cho BK, et al. Prevalence of PER-1 extended-spectrum β-lactamase-clinical isolates of Acinetobacter baumannii in a university hospital, Busan, Korea. Korean J Clin Microbiol. 2004; 7:20–6.
23. De Champs C, Poirel L, Bonnet R, Sirot D, Chanal C, Sirot J, et al. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob Agents Chemother. 2002; 46:3031–4.
24. Naas T, Benaoudia F, Massuard S, Nordmann P. Integron-located VEB-1 extended-spectrum β-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J Antimicrob Chemother. 2000; 46:703–11.
25. Park JH, Lee SH, Jeong SH, Kim BN, Kim KB, Yoon JD, et al. Characterization and prevalence of Escherichia coli and Klebsiella pneumoniae isolates producing an extended spectrum β-lactamase from Korean hospitals. Korean J Lab Med. 2003; 23:18–24.
26. Jeon BC, Jeong SH, Bae IK, Kwon SB, Lee K, Young D, et al. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23-β-lactamase in Korea. J Clin Microbiol. 2005; 43:2241–5.
27. Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex realtime PCR assay and melt curve analysis. J Clin Microbiol. 2007; 45:544–7.

28. Pournaras S, Markogiannakis A, Ikonomidis A, Kondyli L, Bethimouti K, Maniatis AN, et al. Outbreak of multiple clones of imipenem-resistant Acinetobacter baumannii isolates expressing OXA-58 carbapenemase in an intensive care unit. J Antimicrob Chemother. 2006; 57:557–61.
29. Heritier C, Dubouix A, Poirel L, Marty N, Nordmann P. A nosocomial outbreak of Acinetobacter baumannii isolates expressing the carbapenem-hydrolysing oxacillinase OXA-58. J Anti-microb Chemother. 2005; 55:115–8.
30. Lévesque C, Piché L, Larose C, Roy PH. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995; 39:185–91.

31. Shannon KP, French GL. Increasing resistance to antimicrobial agents of Gram-negative organisms isolated at a London teaching hospital, 1995-2000. J Antimicrob Chemother. 2004; 53:818–25.

32. Lee K, Park KH, Jeong SH, Lim HS, Shin JH, Yong D, et al. KONSAR group. Further increase of vancomycin-resistant Enterococcus faecium, amikacin- and fluroquinolone-resistant Klebsiella pneumoniae, and imipenem-resistant Acinetobacter spp. in Korea: 2003 KONSAR surveillance. Yonsei Med J. 2006; 47:43–54.
33. Lee K, Chong Y, Shin HB, Yong D. Rapid increase of imipenem-hydrolyzing Pseudomonas aeruginosa in a Korean hospital. Abstr E-85, 38th ICAAC,. 1998.
34. Lee K, Lee WG, Uh Y, Ha GY, Cho J, Chong Y, et al. VIM- and IMP-type metallo-β-lactamase-producing Pseudomonas spp. and Acinetobacter spp. in Korean hospitals. Emerg Infect Dis. 2003; 9:868–71.
35. Kim IS, Lee NY, Ki CS, Oh WS, Peck KR, Song JH. Increasing prevalence of imipenem-resistant Pseudomonas aeruginosa and molecular typing of metallo-β-lactamase producers in a Korean hospital. Microb Drug Resist. 2005; 11:355–9.
36. Jang SJ. The role of integrons in the spread of multidrug resistance. Korean J Clin Microbiol. 2005; 8:1–9.
37. Shevchenko O, Edelstein M, Kretchikov V, Stratchounski L. Characterization of class 1 integrons carrying the genes for VIM-and IMP-type metallo-β-lactamases in Pseudomonas aeruginosa strains from Russia. Abstr P-1407, 16th ESCMID,. 2006.
38. Jeong SH, Lee K, Chong Y, Yum JH, Lee SH, Choi HJ, et al. Characterization of a new integron containing VIM-2, a metallo-beta-lactamase gene cassette, in a clinical isolate of Enterobacter cloacae. J Antimicrob Chemother. 2003; 51:397–400.
39. Ozgumus OB, Caylan R, Tosun I, Sandalli C, Aydin K, Koksal I. Molecular epidemiology of clinical Pseudomonas aeruginosa isolates carrying IMP-1 metallo-β-lactamase gene in a University Hospital in Turkey. Microb Drug Resist. 2007; 13:191–8.
Go to : 
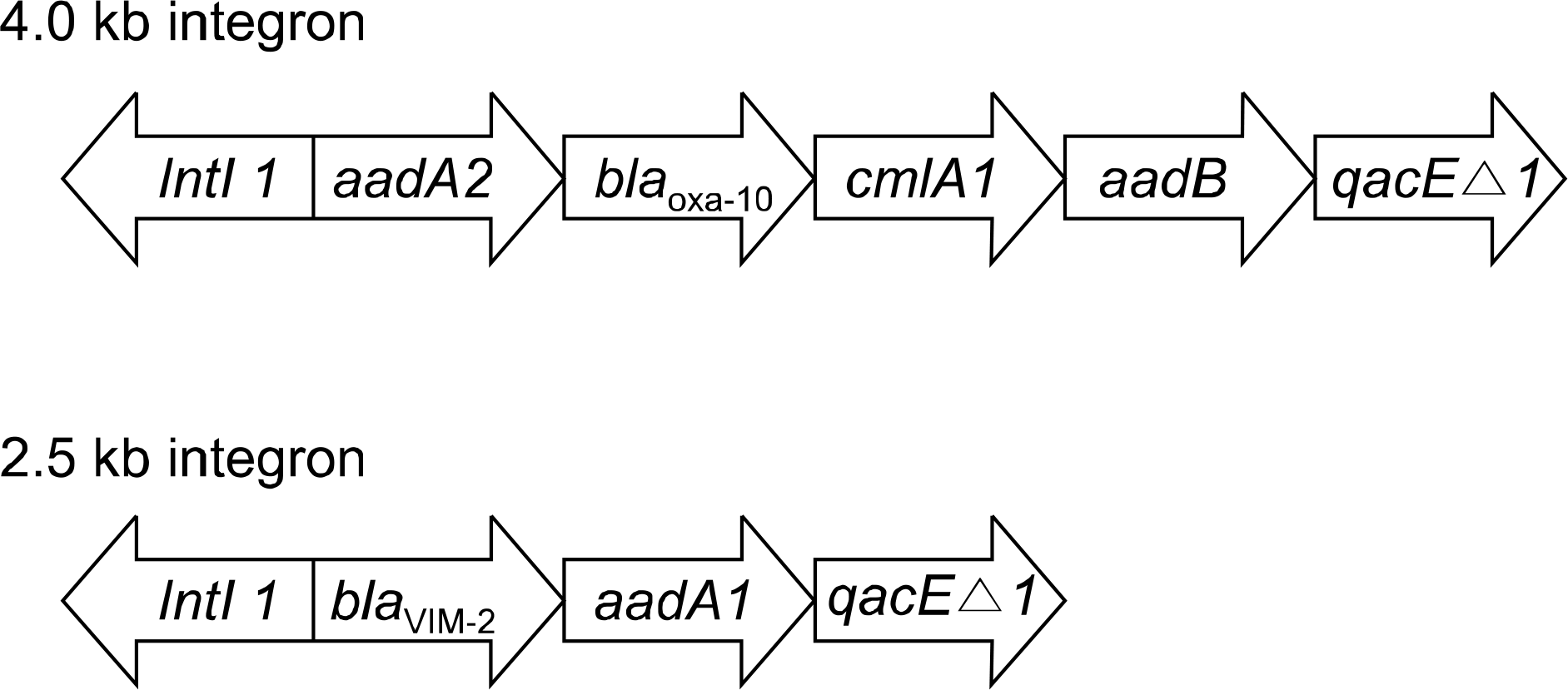 | Fig. 1.Schematic representation of gene cassette structure located in the class 1 integron isolated from Pseudomonas aeruginosa. The horizontal arrows indicate the translation orientation of the genes. |
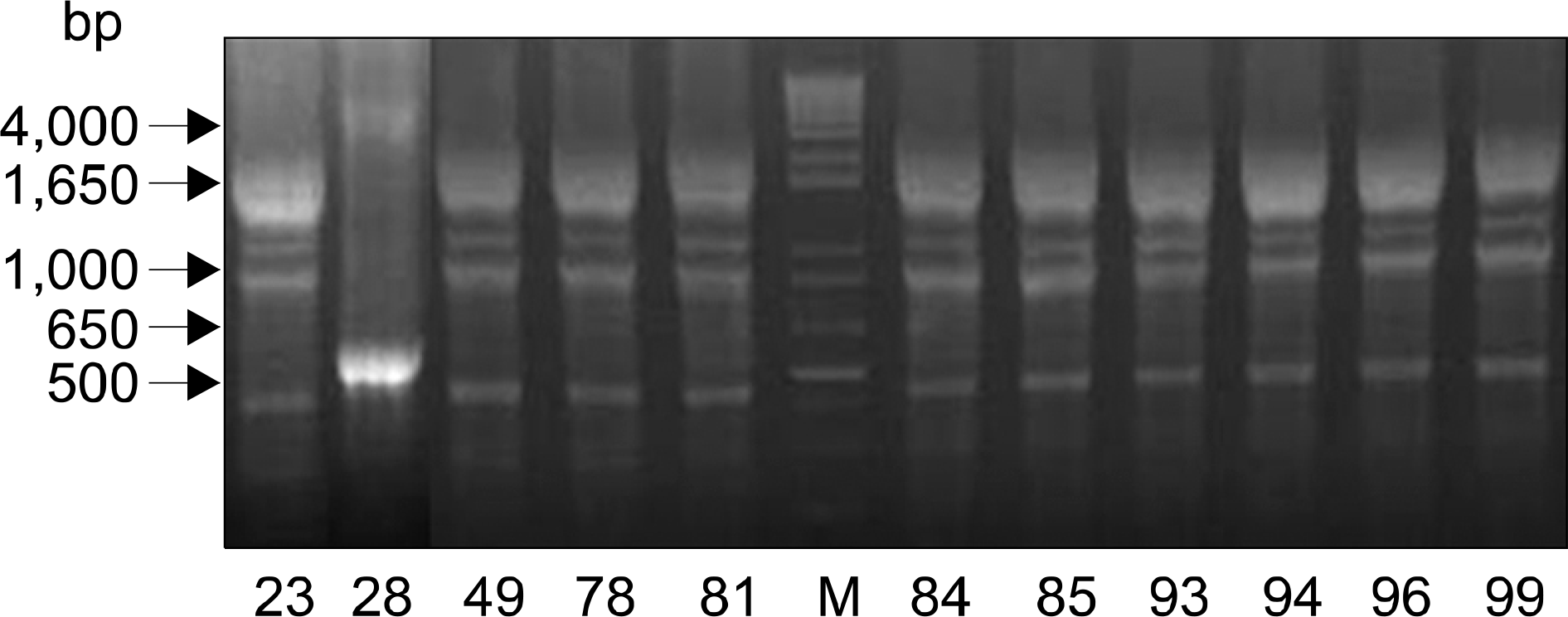 | Fig. 2.Repetitive element sequence-based (Rep)-PCR patterns of genomic DNA from eleven MBL producing Pseudomonas aeruginosa. Lane M is 1 kb DNA size marker. |
Table 1.
Oligonucleotide primers for amplification and sequencing
Table 2.
Antimicrobial susceptibilities of Pseudomonas aeruginosa isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download