Abstract
Background
Blood culture is the definitive method for the diagnosis and treatment of bacteremia and fungemia. Analysis of blood cultures positive for pathogenic species and trends in antimicrobial susceptibility can help delineate appropriate and experimental treatment strategies. In this study, we investigated the incidence of pathogenic species and trends in antimicrobial susceptibility in blood cultures collected from 2003 to 2007 to help clinicians to determine the best methods of diagnosis and treatment. Changes between previously published analyses and this study were also investigated.
Methods
Five-year blood culture results obtained at Kyung Hee University Hospital between 2003 and 2007 were analyzed to determine the bacterial and fungal species present and the antimicrobial susceptibility of the isolates. Antimicrobial susceptibility was tested by the broth microdilution method and the CLSI disk diffusion method.
Results
Among the 66,437 blood cultures, 5,645 were positive. Of the positive blood cultures, 59.8% were positive for aerobic and facultative anaerobic gram-positive cocci. Coagulase-negative staphylococci (CoNS) were frequently isolated. The numbers of anaerobic species and fungi decreased over the years.
Conclusion
CoNS were the microorganisms most commonly isolated from blood cultures at Kyung Hee University Hospital. The number of cultures positive for fungi was higher than that reported in previous studies, but the absolute isolation rate over five years decreased. Anaerobic species were much less frequently isolated than reported for other hospitals.
Go to : 
REFERENCES
1. Trenholme GM, Kaplan RL, Karakusis PH, Stine T, Fuhrer J, Landau W, et al. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J Clin Microbiol. 1989; 27:1342–5.

3. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial disk susceptibility tests; approved standard. 9th ed.CLSI document M2-A9.Wayne, PA: CLSI;2006.
4. Souvenir D, Anderson DE Jr, Palpant S, Mroch H, Askin S, Anderson J, et al. Blood cultures positive for coagulase-negative Staphylococci: antisepsis, pseudobacteremia and therapy of patients. J Clin Microbiol. 1998; 36:1923–6.
5. Koh EM, Lee SG, Kim CK, Kim MS, Yong DE, Lee KW, et al. Microorganisms isolated from blood cultures and their antimicrobial susceptibility patterns at a university hospital during 1994-2003. Korean J Lab Med. 2007; 27:265–75.

6. Chae MJ, Shin JH, Cho DC, Kee SJ, Kim SH, Shin MG, et al. Antifungal susceptibilities and distribution of Candida species recovered from blood cultures over an 8-year period. Korean J Lab Med. 2003; 23:329–35.
7. Ahn GY, Jang SJ, Lee SH, Jeong OY, Chaulagain BP, Moon DS, et al. Trends of the species and antimicrobial susceptibility of microorganisms isolated from blood cultures of patients. Korean J Clin Microbiol. 2006; 9:42–50.
8. Colombo AL, Nucci M, Park BJ, Nouér SA, Artington-Skaggs B, da Matta DA, et al. Epidemiology of candidemia in brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J Clin Microbiol. 2006; 44:2816–23.

9. Marchetti O, Bille J, Fluckiger U, Eggimann P, Ruef C, Garbino J, et al. Epidemiology of candidemia in Swiss tertiary care hospitals: secular trends, 1991-2000. Clin Infect Dis. 2004; 38:311–20.

10. Lee K, Lim CH, Cho JH, Lee WG, Uh Y, Kim HJ, et al. High prevalence of ceftazidime-resistant Klebsiella pneumoniae and increase of imipenem-resistant Pseudomonas aeruginosa and Acinetobacter spp. In Korea: a KONSAR program in 2004. Yonsei Med J. 2006; 47:634–45.
11. Kim ES, Song JS, Lee HJ, Choe PG, Park KH, Cho JH, et al. A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J Antimicrob Chemother. 2007; 60:1108–14.
12. Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997; 40:135–6.
13. Smith TL, Pearson ML, Wilcox KR, Cruz C, Lancaster MV, Robinson-Dunn B, et al. Emergence of vancomycin resistance in Staphylococcus aureus. Glycopiptide-intermediate Staphylococcus aureus working group. N Engl J Med. 1999; 340:493–501.
14. Fridkin SK. Vancomycin-intermediate and resistant Staphylococcus aureus: what the infectious disease specialist needs to know. Clin Infect Dis. 2001; 32:108–15.
Go to : 
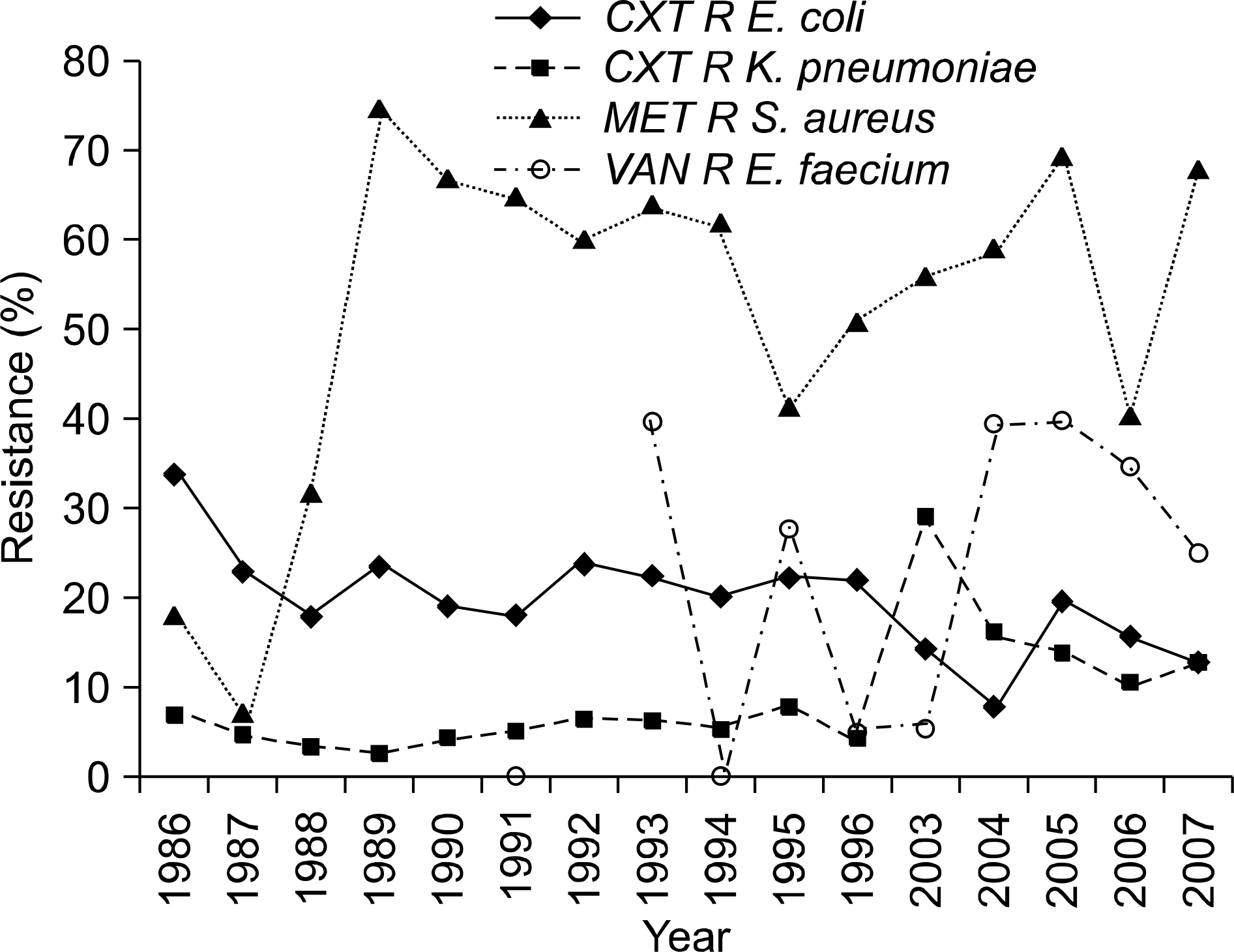 | Fig. 1.Trends in antimicrobial resistances (%) of E. coli, K. pneumoniae, S. aureus and E. faecium. Abbreviations: CXT R, cefotaxim-resistant; MET R, methicillin- resistant; VAN R, vancomycin-resistant. |
Table 1.
Groups of bacteria and fungi isolated by year
Table 2.
Annual isolation of significant bacterial pathogens
Table 3.
Species of fungi isolated




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download