Abstract
Background
Direct immunofluorescence assay (DFA) and shell vial culture (SVC) have been used to diagnose respiratory viral infections. Recently a multiplex reverse transcriptase PCR (mRT-PCR) for 12 respiratory viruses has been introduced. We evaluated the diagnostic usefulness of these methods.
Methods
Among 275 nasopharyngeal aspirates (NPAs) received from pediatric patients during the 3-month period from May through July, 2007, 122 samples were selected so as to include diverse viruses and varying numbers of DFA-positive cells for mRT-PCR. Also, the results of the 85 NPAs that had been analyzed by both DFA and SVC were reviewed retrospectively.
Results
Detection rates for the seven major respiratory viruses, respiratory syncytial virus (RSV), influenza virus A and B, parainfluenza virus 1, 2, and 3, and adenovirus by DFA vs mRT-PCR were 32.0% and 55.7%, and by DFA vs SVC were 32.9% and 40.0%. A number of adenovirus detected by DFA vs mRT-PCR were 12 and 22, and by DFA vs SVC were 6 and 18. A number of RSV detected were 3 and 6, and 13 and 8, respectively.
Go to : 
REFERENCES
1. Henrickson KJ, Hoover S, Kehl KS, Hua W. National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr Infect Dis J. 2004; 23:11S–8S.

2. Larson HE, Reed SE, Tyrrell DA. Isolation of rhinoviruses and coronaviruses from 38 colds in adults. J Med Virol. 1980; 5:221–9.

3. Kwak YH, Choi EH, Lee HJ. Detection of rhinovirus from children with lower respiratory tract infections by reverse transcription polymerase chain reaction. Infect Chemother. 2003; 49:1–11.
4. Chung JY, Han TH, Kim SW, Hwang ES. Respiratory picornavirus infections in Korean children with lower respiratory tract infections. Scand J Infect Dis. 2007; 39:250–4.

5. van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001; 7:719–24.

6. Ogilvie M. Molecular techniques should not now replace cell culture in diagnostic virology laboratories. Rev Med Virol. 2001; 11:351–4.
7. Carman B. Molecular techniques should now replace cell culture in diagnostic virology laboratories. Rev Med Virol. 2001; 11:347–9.

8. Niesters HG. Molecular and diagnostic clinical virology in real time. Clin Microbiol Infect. 2004; 10:5–11.

9. Roh KH, Kim J, Nam MH, Yoon S, Lee CK, Lee K, et al. Comparison of the Seeplex reverse transcription PCR assay with the R-mix viral culture and immunofluorescence techniques for detection of eight respiratory viruses. Ann Clin Lab Sci. 2008; 38:41–6.
10. Jennings LC, Anderson TP, Werno AM, Beynon KA, Murdoch DR. Viral etiology of acute respiratory tract infections in children presenting to hospital: role of polymerase chain reaction and demonstration of multiple infections. Pediatr Infect Dis J. 2004; 23:1003–7.
11. Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007; 20:49–78.

12. Arnold JC, Singh KK, Spector SA, Sawyer MH. Undiagnosed respiratory viruses in children. Pediatrics. 2008; 121:e631–7.

13. Kang JO, Kim EC, Lee KM, Lee NY, Lee CK. Surveillance for respiratory virus testing situation in Korea and epidemiology for the respiratory viruses detected in 5 university hospitals -Report from virus study group. Korean J Clin Microbiol. 2007; 10:102–8.
14. Yoo SJ, Kuak EY, Shin BM. Detection of 12 respiratory viruses with two-set multiplex reverse transcriptase-PCR assay using a dual priming oligonucleotide system. Korean J Lab Med. 2007; 27:420–7.

15. Matthey S, Nicholson D, Ruhs S, Alden B, Knock M, Schultz K, et al. Rapid detection of respiratory viruses by shell vial culture and direct staining by using pooled and individual monoclonal antibodies. J Clin Microbiol. 1992; 30:540–4.

16. Landry ML, Ferguson D. SimulFluor respiratory screen for rapid detection of multiple respiratory viruses in clinical specimens by immunofluorescence staining. J Clin Microbiol. 2000; 38:708–11.

17. Dunn JJ, Woolstenhulme RD, Langer J, Carroll KC. Sensitivity of Respiratory virus culture when screening with R-mix fresh cells. J Clin Microbiol. 2004; 42:79–82.

18. Hindiyeh M, Hillyard DR, Carroll KC. Evaluation of the Prodesse Hexaplex multiplex PCR assay for direct detection of seven respiratory viruses in clinical specimens. Am J Clin Pathol. 2001; 116:218–24.

19. Kehl SC, Henrickson KJ, Hua W, Fan J. Evaluation of the Hexaplex assay for detection of respiratory viruses in children. J Clin Microbiol. 2001; 39:1696–701.

20. Freymuth F, Vabret A, Cuvillon-Nimal D, Simon S, Dina J, Legrand L, et al. Comparison of multiplex PCR assays and conventional techniques for the diagnostic of respiratory virus infections in children admitted to hospital with an acute respiratory illness. J Med Virol. 2006; 78:1498–504.

21. Chun JY, Kim KJ, Hwang IT, Kim YJ, Lee DH, Lee IK, et al. Dual priming oligonucleotide system for the multiplex detection of respiratory viruses and SNP genotyping of CYP2C19 gene. Nucleic Acids Res. 2007; 35:e40.

22. Sung H, Park SJ, Woo YD, Choi BH, Kim MN. Evaluation of Seeplex RV detection kit for detecting rhinovirus, human meta-pneumovirus and coronavirus. Korean J Lab Med. 2008; 28:109–17.
Go to : 
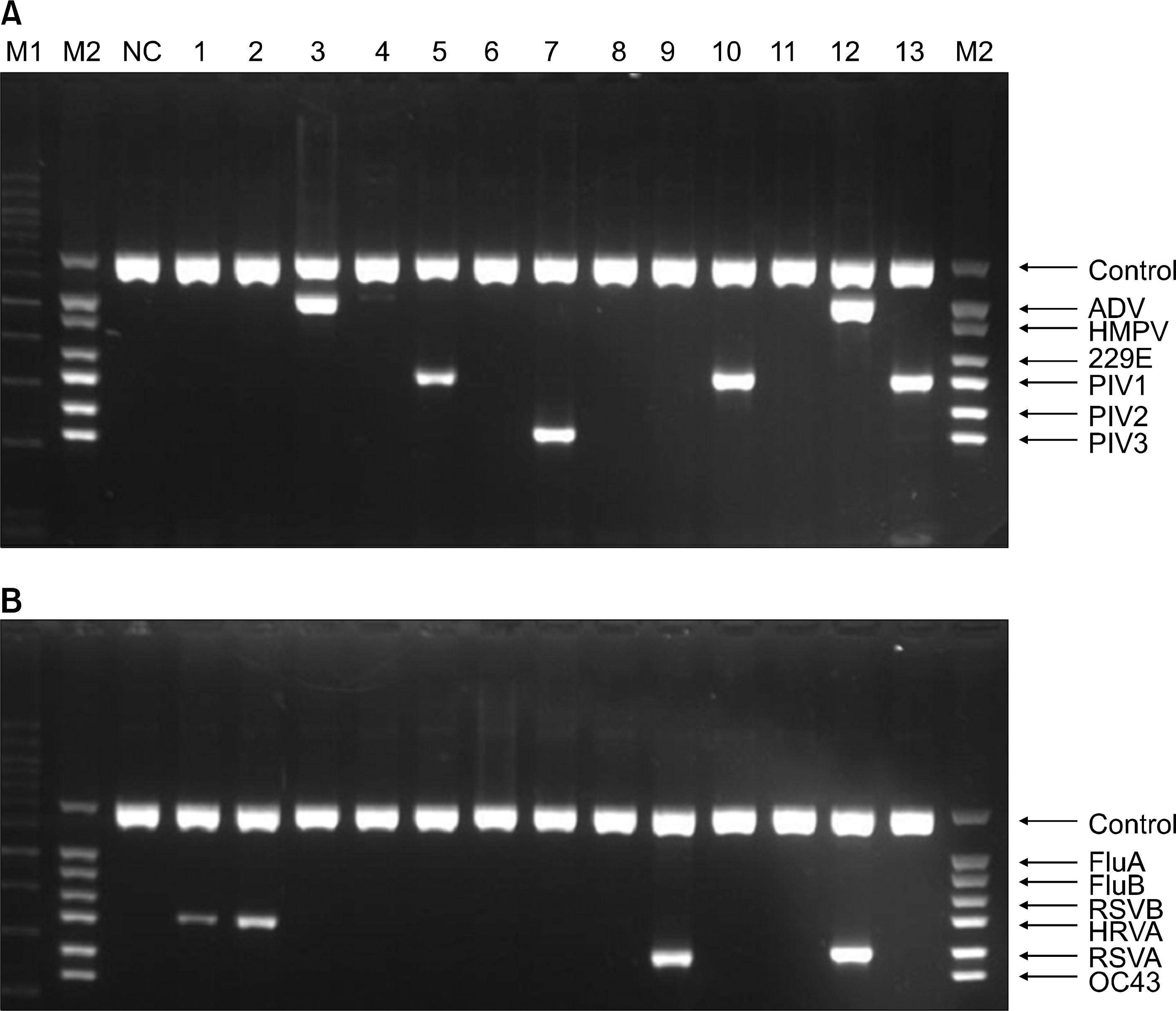 | Fig. 1.Electrophoresis of mRT-PCR products for 12 repiratory viruses. Abbreviations: Control, internal control; ADV, adenovirus; HMPV, human metapneumo-virus; 229E, coronavirus 229E/NL63; PIV1, PIV2, PIV3, parainfluenza virus 1, 2, 3; FluA, FluB, influenza virus A, B; RSVA, B, respiratory syncytial virus A, B; HRVA, human rhinovirus A; OC43, coronavirus OC43. M1, 100 bp size marker; M2, viruses marker; NC, negative control; lane 1∼13, patients: patient 1 HRVA, patient 2 HRVA, patient 3 ADV, patient 5 PIV1, patient 7 PIV3, patient 9 RSVA, patient 10 PIV1, patient 12 ADV+RSVA, and patient 13 PIV1. Other patients were negative. |
Table 1.
Detection of 7 major respiratory viruses in nasopharyngeal aspirates (n=122) by DFA and mRT-PCR methods
| DFA | mRT-PCR | |
|---|---|---|
| ADV | 11 | 20 (PIV3)∗ |
| FluA | 10 | 12 (ADV)∗ |
| FluB | 1 | 4 |
| PIV1 | 5 | 8 (PIV3)∗ |
| PIV2 | 0 | 1 |
| PIV3 | 9 | 17 (PIV2)∗ |
| RSV | 3 (ADV)∗ | 6 (ADV)∗ |
| Negative | 83 | 54 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download