Abstract
Background
Recently, there have been reports of in-fections with multidrug-resistant Pseudomonas aeruginosa. To determine the mechanism of the resistance, we investigated the prevalence of Ambler class A and D β-lactamases, their extended-spectrum derivatives, and class B and D carbapenemase in mul-tidrug-resistant P. aeruginosa isolates.
Methods
During the period of March 2006 to May 2007, clinical isolates of multidrug-resistant P. aeruginosa were collected from patients in Chungnam National University Hospital, Daejeon, Korea. Inhibitor-potentiated disk diffusion tests were used for the screening of metallo-β-lactamase (MBL) production. PCR and DNA sequencing were conducted for the detection of β-lactamase genes. We also employed the enterobacterial repetitive intergenic consensus (ERIC)-PCR method for an epidemiologic study.
Results
A total of 37 consecutive, non-duplicate, multidrug-resistant P. aeruginosa were isolated. Twentynine of 37 isolates harbored blaOXA-10 (56.8%), blaOXA-2 (18.9%), and blaOXA-1 (5.4%). Only one iso-late produced IMP-1, and it also harbored blaOXA-1. None harbored Ambler class A β-lactamase or class D carbapenemase. The strains producing OXA type β-lactamases showed a significantly higher resistance to aminoglycoside compared to non-producers. The ERIC-PCR pattern of the 19 OXA-10 producing strains indicated that the isolates were closely related in terms of clonality.
Conclusion
OXA type β-lactamases are the most prevalent among the acquired β-lactamases produced by multidrug-resistant P. aeruginosa isolated at a university hospital in Chungcheong Province. Besides β-lactam antibiotics, the strains harboring OXA type β-lactamase showed a significantly higher resistance to aminoglycoside and qunolone.
Go to : 
References
1. Bergogne-Berezin E. Pseudomonas and miscellaneous gram-negative bacilli. Cohen J and Powerly WG, editor. Infectious Diseases. 2nd ed.New York: Mosby;2003. p. 2203–17.
2. Song W, Woo HJ, Kim JS, Lee KM. In vitro activity of β-lactams in combination with other antimicrobial agents against resistant strains of Pseudomonas aeruginosa. Int J Antimicrob Agents. 2003; 21:8–12.

3. Lee K, Park KH, Jeong SH, Lim HS, Shin JH, Yong D, et al. KONSAR group. Further increase of vancomycin-resistant Enterococcus faecium, amikacin- and fluroquinolone-resistant Klebsiella pneumoniae, and imipenem-resistant Acinetobacter spp. in Korea: 2003 KONSAR surveillance. Yonsei Med J. 2006; 47:43–54.
4. McGowan JE Jr. Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum. Am J Infect Control. 2006; 34:S29–S37.

5. Weldhagen GF, Poirel L, Nordmann P. Ambler class A extended spectrum β-lactamases in Pseudomonas aeruginosa: novel developments and clinical impact. Antimicrob Agents Chemother. 2003; 47:2385–92.
6. Poirel L, Naas T, Guibert M, Chaibi EB, Labia R, Nordmann P. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob Agents Chemother. 1999; 43:573–81.
7. Nordmann P, Ronco E, Naas T, Duport C, Michel-Briand Y, Labia R. Characterization of a novel extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1993; 37:962–9.
8. Chen HY, Yuan M, Livemore DM. Mechanisms of resistance to β-lactam antibiotics amongst Pseudomonas aeruginosa isolates collected in the UK in 1993. J Med Microbiol. 1995; 43:300–9.

9. Lee S, Park YJ, Kim M, Lee HK, Han K, Kang CS, et al. Prevalence of Ambler class A and D β-lactamases among clinical isolates of Pseudomonas aeruginosa in Korea. J Antimicrob Chemother. 2005; 56:122–7.

10. Poirel L, Weldhagen GF, Naas T, De Champs C, Dove MG, Nordmann P. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob Agents Chemother. 2001; 45:2598–603.
11. Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991; 35:147–51.

12. Riccio ML, Franceschini N, Boschi L, Caravelli B, Cornaglia G, Fontana R, et al. Characterization of the metallo-β-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of blaIMP allelic variants carried by gene cassettes of different phylogeny. Antimicrob Agents Chemother. 2000; 44:1229–35.
13. Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, et al. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999; 43:1584–90.
14. Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, et al. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob Agents Chemother. 2002; 46:1053–8.
15. Toleman MA, Simm AM, Murphy TA, Gales AC, Biedenbach DJ, Jones RN, et al. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J Antimicrob Chemother. 2002; 50:673–9.
16. Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob Agents Chemother. 2004; 48:4654–61.
17. Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier JD, et al. Novel acquired metallo-β-lactamase gene, bla(SIM-1), in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother. 2005; 49:4485–91.
18. Donald HM, Scaife W, Amyes SG, Young HK. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob Agents Chemother. 2000; 44:196–9.
19. Bou G, Oliver A, Martinez-Beltran J. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob Agents Chemother. 2000; 44:1556–61.
20. Yoon WS, Lee BY, Bae IK, Kwon SB, Jeong SH, Jeong TJ, et al. Prevalence of imipenem-resistant Pseudomonas aeruginosa isolates and mechanisms of resistance. Korean J Clin Microbiol. 2005; 8:26–33.
21. Timurkaynak F, Can F, Azap OK, Demirbilek M, Arslan H, Karaman SO. In vitro activities of non-traditional antimicrobials alone or in combination against multidrug-resistant strains of Pseudomonas aeruginosa and Acinetobacter baumannii isolated from intensive care units. Int J Antimicrob Agents. 2006; 27:224–8.

22. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. M100-S10 (M2). Wayne, Pensylvania; CLSI. 2006.
23. Oh EJ, Lee S, Park YJ, Park JJ, Park K, Kim SI, et al. Prevalence of metallo-β-lactamase among Pseudomonas aeruginosa and Acinetobacter baumannii in a Korean university hospital and comparison of screening methods for detecting metallo-β-lac-tamse. J Microbiol Methods. 2003; 54:411–8.
24. Kang JH, Bae IK, Kwon SB, Jeong SH, Lee J, Lee WG, et al. Prevalence of ambler class A extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates in Korea. Korean J Clin Microbiol. 2005; 8:17–25.
25. Naas T, Benaoudia F, Massuard S, Nordmann P. Integron-located VEB-1 extended-spectrum β-lactamase gene in a Proteus mirabilis clinical isolate from Vietnam. J Antimicrob Chemother. 2000; 46:703–11.
26. Park JH, Lee SH, Jeong SH, Kim BN, Kim KB, Yoon JD, et al. Characterization and prevalence of Escherichia coli and Klebsiella pneumoniae isolates producing an extended spectrum β-lactamase from Korean hospitals. Korean J Lab Med. 2003; 23:18–24.
27. Jeon BC, Jeong SH, Bae IK, Kwon SB, Lee K, Young D, et al. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23-β-lactamase in Korea. J Clin Microbiol. 2005; 43:2241–5.
28. Aubert D, Poirel L, Chevalier J, Leotard S, Pages JM, Nordmann P. Oxacillinase-mediated resistance to cefepime and susceptibility to ceftazidime in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2001; 45:1615–20.
29. De Champs C, Poirel L, Bonnet R, Sirot D, Chanal C, Sirot J, et al. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob Agents Chemother. 2002; 46:3031–4.
30. Heritier C, Dubouix A, Poirel L, Marty N, Nordmann P. A nosocomial outbreak of Acinetobacter baumannii isolates expressing the carbapenem-hydrolysing oxacillinase OXA-58. J Antimicrob Chemother. 2005; 55:115–8.
31. Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, et al. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol. 2007; 45:544–7.

32. Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991; 19:6823–31.
33. Park KO, Son HC, Bae IK, Jeong SH. Molecular epidemiology of infection caused by OXA-23 or IMP-1 β-lactamase-producing Acinetobacter baumannii. Korean J Lab Med. 2005; 8:121–9.
34. Yao JDC and Moellering RC Jr. Antibacterial agents. Murray PR, Baron EJ, editors. Manual for Clinical Microbiology. 7th ed.Washington: American Society for Microbiology;1999. p. 1474–504.
35. Danel F, Hall LM, Duke B, Gur D, Livermore DM. OXA-17, a further extended-spectrum variant of OXA-10 β-lactamase, isolated from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999; 43:1362–6.
36. Aktaş Z, Poirel L, Salcioğlu M, Ozcan PE, Midilli K, Bal C, et al. PER-1- and OXA-10-like β-lactamases in ceftazidime-resistant Pseudomonas aeruginosa isolates from intensive care unit patients in Istanbul, Turkey. Clin Microbiol Infect. 2005; 11:193–8.

37. Poirel L, Girlich D, Naas T, Nordmann P. OXA-28, an extended-spectrum variant of OXA-10 β-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob Agents Chemother. 2001; 45:447–53.
38. Yan JJ, Tsai SH, Chuang CL, Wu JJ. OXA-type β-lactamases among extended-spectrum cephalosporin-resistant Pseudomonas aeruginosa isolates in a university hospital in southern Taiwan. J Microbiol Immunol Infect. 2006; 39:130–4.
39. Oh SJ, Lee SU, Hwang HY, Bae IK, Jo HS, Lee BH, et al. Prevalence of class a extended-spectrum β-lactamases in clinical isolates of Acinetobacter baumannii and Pseudomonas aeruginosa. Korean J Lab Med. 2006; 26:14–20.

40. Nordmann P and Guibert M. Extended-spectrum β-lactamases in Pseudomonas aeruginosa. J Antimicrob Chemother. 1998; 42:128–31.
41. Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-β-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. 1995; 39:824–9.
42. Ohara M, Kouda S, Onodera M, Fujiue Y, Sasaki M, Kohara T, et al. Molecular characterization of imipenem-resistant Pseudomonas aeruginosa in Hiroshima, Japan. Microbiol Immunol. 2007; 51:271–7.
Go to : 
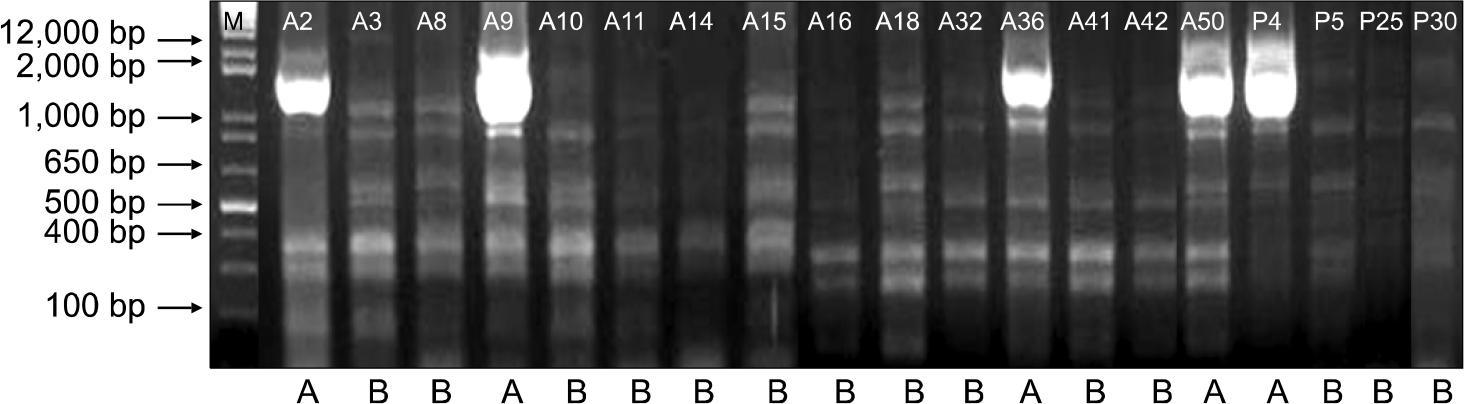 | Fig. 1.ERIC-PCR patterns of genomic DNA from clinical isolates of multi-drug resistant Pseudomonas aeruginosa harboring blaOXA-10. Lane M is 1 kb DNA size marker. Nineteen strains of OXA-10 β-lactamase producing clinical isolates show A or B pattern. |
Table 1.
Oligonucleotides used as primers for amplification and sequencing
Table 2.
Antibiotic susceptability profiles and β-lactamases of multi-drug resistant Pseudomonas aeruginosa
Table 3.
Prevalence of Ambler class A, B, and D β-lactamases in multi-drug resistant Pseudomonas aeruginosa
| Class | Type of β-lactamases | No.(%) of isolates |
|---|---|---|
| Class B | IMP-1 | 1 (2.7)∗7 |
| Class D | OXA-1 | 2 (5.4)∗ |
| OXA-2 | 7 7 (18.9)† | |
| OXA-10 | 20 (54.1)† | |
| Combined | IMP and OXA-1 | 1 (2.7)∗ |
| Combined | OXA-2 and OXA10 | 7 1 (2.7)†7 |
| Not detected | 9 (24.3) |
Table 4.
Comparison of the antimicrobial resistance (%) between the class D β-lactamase producers and non-producers




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download