Abstract
Background
Clinical isolates of AmpC β-lactamase-producing Enterobacteriaceae were evaluated to determine the prevalence of CTX-M extended-spectrum β-lactamases (ESBLs) and their genetic environments.
Methods
A total of 250 non-duplicate isolates of Eneterobacter aerogenes, E. cloacae, Citrobacter freundii, Serratia marcescens and Morganella morganii were collected at a Korean hospital. ESBL production was determined by double disk synergy test. For ESBL producers, bla genes were sequenced and blaCTX-M environment was characterized by PCR mapping and sequencing.
Results
Among the 250 isolates 29 (11.6%) produced ESBL, and 14 of the 29 isolates produced CTX-M ESBLs, including CTX-M-9 by 8 isolates, CTX-M-3 by 4 isolates, CTX-M-12 by 1 isolate, and CTX-M-14 by 1 isolate. ISEcp1 was present upstream of blaCTX-M-3, 12, and 14. Three of the four CTX-M-3 producers had the same genetic environment (pemK-ISEcp1-blaCTX-M-3-orf477-mucA). An IS903-like element was found downstream of blaCTX-M-14. ISCR1 was identified upstream of blaCTX-M-9 and ISCR1 and blaCTX-M-9 were located on sul1-type class 1 integron. The variable region between the 5'-CS and the first 3'-CS contained dfrA16 and aadA2. Its structure was similar to that of In60, but our isolates did not have IS3000 or second 3'-CS.
Go to : 
References
1. Eckert C, Gautier V, Saladin-Allard M, Hidri N, Verdet C, Ould-Hocine Z, et al. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother. 2004; 48:1249–55.
2. Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr Opin Pharmacol. 2007; 7:459–69.

3. Ryoo NH, Kim EC, Hong SG, Park YJ, Lee K, Bae IK, et al. Dissemination of SHV-12 and CTX-M-type extended-spectrum β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J Antimicrob Chemother. 2005; 56:698–702.

4. Kim JY, Park YJ, Kim SI, Kang MW, Lee SO, Lee KY. Nosocomial outbreak by Proteus mirabilis producing extended-spectrum β-lactamase VEB-1 in a Korean university hospital. J Antimicrob Chemother. 2004; 54:1144–7.

5. Walther-Rasmussen J and H⊘iby N. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum β-lactamases. Can J Microbiol. 2004; 50:137–65.
6. Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004; 48:1–14.

7. Eckert C, Gautier V, Arlet G. DNA sequence analysis of the genetic environment of various blaCTX-M genes. J Antimicrob Chemother. 2006; 57:14–23.
8. Brizio A, Vasco S, Goncalves AR, Lito LM, Cristino JM, Salgado MJ, et al. Survey of extended-spectrum β-lactamases in Escherichia coli isolates from a Portuguese hospital and characterisation of a novel class 1 integron (In60A) carrying the blaCTX-M-9 gene. Int J Antimicrob Agents. 2006; 28:320–4.
9. Canton R and Coque TM. The CTX-M β-lactamase pandemic. Curr Opin Microbiol. 2006; 9:466–75.
10. Shibata N, Kurokawa H, Doi Y, Yagi T, Yamane K, Wachino J, et al. PCR classification of CTX-M-type β-lactamase genes identified in clinically isolated gram-negative bacilli in Japan. Antimicrob Agents Chemother. 2006; 50:791–5.

11. Pai H, Choi EH, Lee HJ, Hong JY, Jacoby GA. Identification of CTX-M-14 extended-spectrum β-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J Clin Microbiol. 2001; 39:3747–9.
12. Bae IK, Jeong SH, Lee K, Yong D, Lee J, Hong SG, et al. Emergence of CTX-M-12 and a novel CTX-M type extended-spectrum β-lactamaseproducing Klebsiella pneumoniae. Korean J Lab Med. 2006; 26:21–6.
13. Kim J, Lim YM, Jeong YS, Seol SY. Occurrence of CTX-M-3, CTX-M-15, CTX-M-14, and CTX-M-9 extended-spectrum β-lactamases in Enterobacteriaceae clinical isolates in Korea. Antimicrob Agents Chemother. 2005; 49:1572–5.
14. Kim YR, Kim SI, Lee JY, Park YJ, Lee KY, Kang MW. Nosocomial transmission of CTX-M-15 and OXA-30 β-lactamase-producing Escherichia coli in a neurosurgical intensive care unit. Ann Clin Lab Sci. 2005; 35:297–301.
15. Oh CE, Hong JS, Bae IK, Song EH, Jeong SH, Lee KW, et al. Dissemination of CTX-M type extended-spectrum β-lactamases and emergence of CTX-M-12 in Escherichia coli. Korean J Lab Med. 2005; 25:252–8.
16. Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Disk Susceptibility Tests-Ninth Edition: Approved Standard M2-A9. Wayne, PA; CLSI. 2006.
17. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995; 33:2233–9.

18. Winokur PL, Canton R, Casellas JM, Legakis N. Variations in the prevalence of strains expressing an extended-spectrum β-lactamase phenotype and characterization of isolates from Europe, the Americas, and the Western Pacific region. Clin Infect Dis. 2001; 32(Suppl 2):S94–103.

19. Lee JH, Bae IK, Kwon SB, Jeong SH, Woo GJ, Lee J, et al. Prevalence of CTX-M-type extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae Isolates in Korea, 2003. Korean J Clin Microbiol. 2004; 7:111–8.
20. Park JH, Lee SH, Jeong SH, Kim BN, Kim KB, Yoon JD, et al. Characterization and prevalence of Escherichia coli and Klebsiella pneumoniae isolates producing an extended-spectrum β-lactamase from Korean hospitals. Korean J Lab Med. 2003; 23:18–24.
21. Tzelepi E, Giakkoupi P, Sofianou D, Loukova V, Kemeroglou A, Tsakris A. Detection of extended-spectrum β-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol. 2000; 38:542–6.
22. Park YJ, Park SY, Oh EJ, Park JJ, Lee KY, Woo GJ, et al. Occurrence of extended-spectrum β-lactamases among chromosomal AmpC-producing Enterobacter cloacae, Citrobacter freundii, and Serratia marcescens in Korea and investigation of screening criteria. Diagn Microbiol Infect Dis. 2005; 51:265–9.

23. Hoffmann H, Sturenburg E, Heesemann J, Roggenkamp A. Prevalence of extended-spectrum β-lactamases in isolates of the Enterobacter cloacae complex from German hospitals. Clin Microbiol Infect. 2006; 12:322–30.

24. Ho PL, Shek RH, Chow KH, Duan RS, Mak GC, Lai EL, et al. Detection and characterization of extended-spectrum β-lactamases among bloodstream isolates of Enterobacter spp. in Hong Kong, 2000–2002. J Antimicrob Chemother. 2005; 55:326–32.

25. Yu WL, Cheng KC, Chi CJ, Chen HE, Chuang YC, Wu LT. Characterisation and molecular epidemiology of extended-spectrum β-lactamase-producing Enterobacter cloacae isolated from a district teaching hospital in Taiwan. Clin Microbiol Infect. 2006; 12:579–82.

26. Pitout JD, Reisbig MD, Venter EC, Church DL, Hanson ND. Modification of the double-disk test for detection of Enterobacteriaceae producing extended-spectrum and AmpC β-lactamases. J Clin Microbiol. 2003; 41:3933–5.
27. Bae IK, Lee BH, Hwang HY, Jeong SH, Hong SG, Chang CL, et al. A novel ceftazidime-hydrolysing extended-spectrum β-lactamase, CTX-M-54, with a single amino acid substitution at position 167 in the omega loop. J Antimicrob Chemother. 2006; 58:315–9.

28. Jeong SH, Bae IK, Kwon SB, Lee JH, Song JS, Jung HI, et al. Dissemination of transferable CTX-M-type extended-spectrum β-lactamase-producing Escherichia coli in Korea. J Appl Microbiol. 2005; 98:921–7.
29. Jeong SH, Bae IK, Lee JH, Sohn SG, Kang GH, Jeon GJ, et al. Molecular characterization of extended-spectrum β-lactamases produced by clinical isolates of Klebsiella pneumoniae and Escherichia coli from a Korean nationwide survey. J Clin Microbiol. 2004; 42:2902–6.
30. Pai H. The characteristics of extended-spectrum β-lactamases in Korean isolates of Enterobacteriaceae. Yonsei Med J. 1998; 39:514–9.
31. Pai H, Lyu S, Lee JH, Kim J, Kwon Y, Kim JW, et al. Survey of extended-spectrum β-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J Clin Microbiol. 1999; 37:1758–63.
32. Kariuki S, Corkill JE, Revathi G, Musoke R, Hart CA. Molecular characterization of a novel plasmid-encoded cefotaximase (CTX-M-12) found in clinical Klebsiella pneumoniae isolates from Kenya. Antimicrob Agents Chemother. 2001; 45:2141–3.
33. Lartigue MF, Poirel L, Nordmann P. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol Lett. 2004; 234:201–7.
34. Golebiewski M, Kern-Zdanowicz I, Zienkiewicz M, Adamczyk M, Zylinska J, Baraniak A, et al. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob Agents Chemother. 2007; 51:3789–95.
35. Poirel L, Lartigue MF, Decousser JW, Nordmann P. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother. 2005; 49:447–50.
36. Novais A, Canton R, Valverde A, Machado E, Galan JC, Peixe L, et al. Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-α, and IncFI groups. Antimicrob Agents Chemother. 2006; 50:2741–50.
37. Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, et al. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in longterm-care facilities in Toronto, Canada. Antimicrob Agents Chemother. 2004; 48:3758–64.
38. Morosini MI, Garcia-Castillo M, Coque TM, Valverde A, Novais A, Loza E, et al. Antibiotic coresistance in extended-spectrum-β-lactamase-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob Agents Chemother. 2006; 50:2695–9.
39. Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, et al. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. 2006; 50:1178–82.
40. Nordmann P and Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother. 2005; 56:463–9.
Go to : 
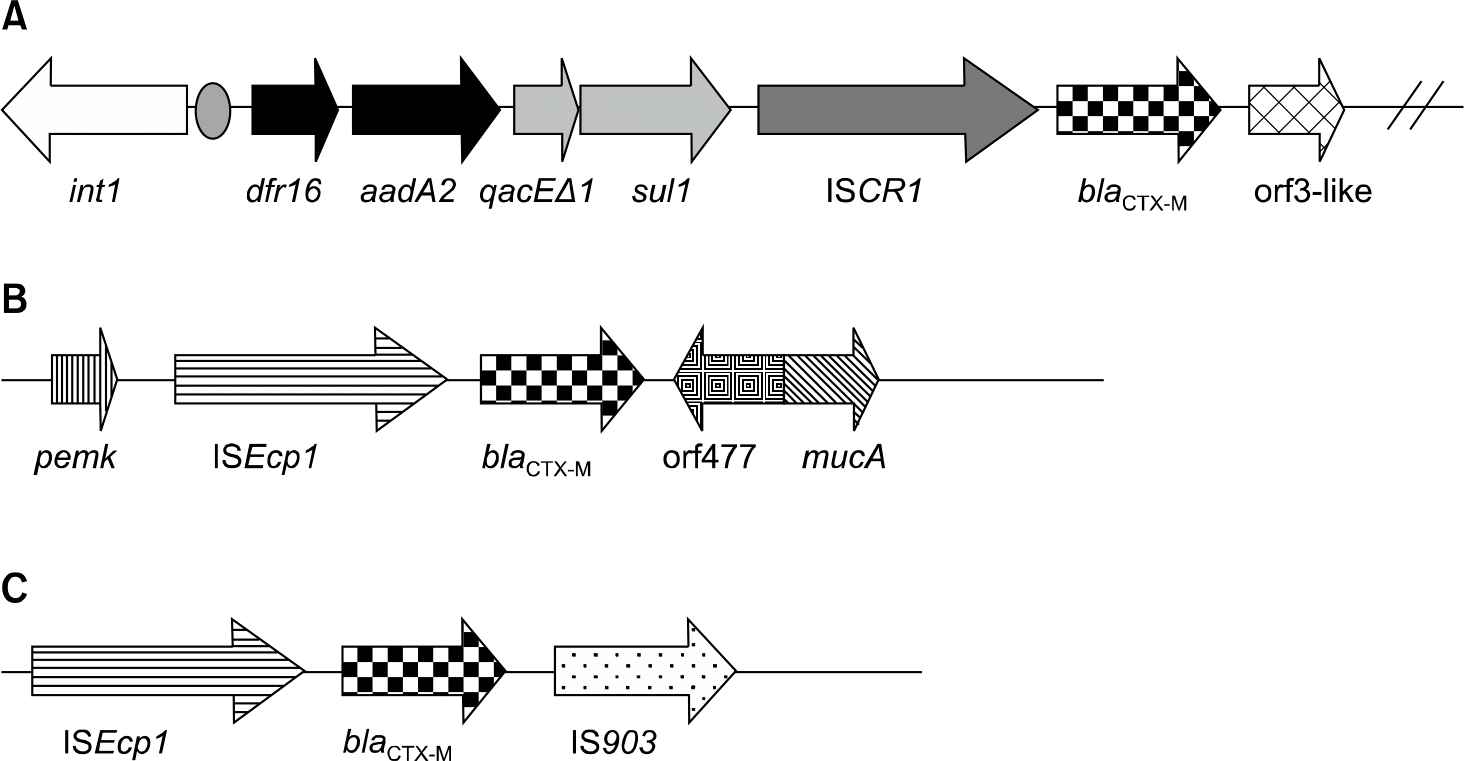 | Fig. 1.Schematic representation of the genetic environment of blaCTX-M genes. (A) C. freundii 06–5-U1734 and E. cloacae 06–2-R1363 (CTX-M-9), (B) C. freundii 06–5-P448, S. marcescens 06-6-R1088 and 06-7-B4288 (CTX-M-3), (C) M. morganii 06-8-P334 (CTX-M-14). |
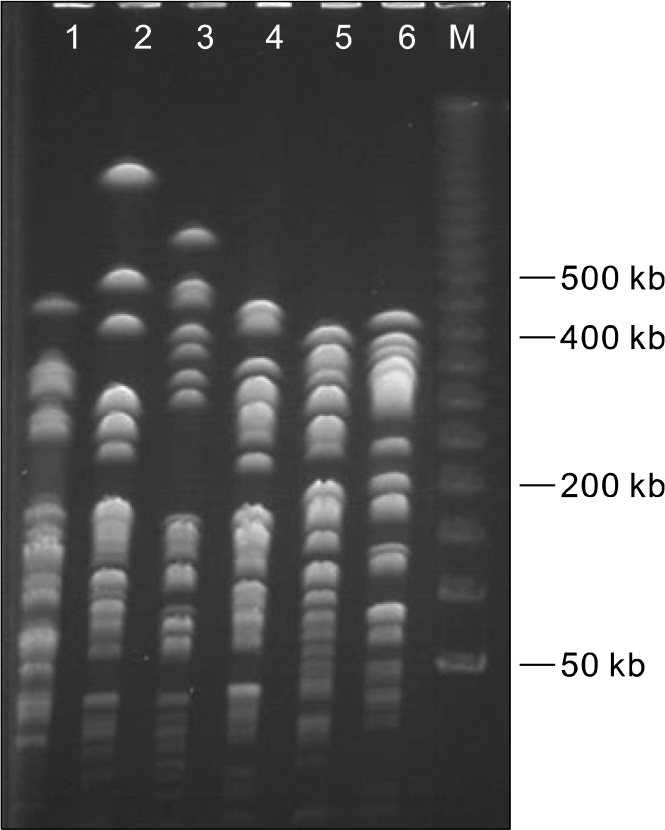 | Fig. 2.PFGE analysis of CTX-M-9 producing E. cloacae isolates. Lane 1, 06-2-R1363; Lane 2, 06-5-R1137; Lane 3, 06-5-R468; Lane 4, 06–5-C242; Lane 5, 06–5-R1186; Lane 6, 06–5-R1124. Lane; M, DNA molecular marker. |
Table 1.
Sequences of the primers for bla genes and blaCTX-M genetic environment
Table 2.
Types of ESBL produced by species
Table 3.
Characteristics of CTX-M type ESBLs in clinical isolates




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download