Abstract
Background
Clarithromycin resistance in Helicobacter pylori is a major cause of eradication therapy failure. The objective of this study was to determine the frequency and type of mutations in the 23S rRNA gene in Korea, which are associated with clarithromycin resistance.
Methods
From January 2008 to March 2008, 353 gastric biopsy specimens were collected from five university hospitals in Seoul and Kyunggido. H. pylori infection was defined as showing a positive result in at least one of the following three tests: a microaerophilic culture, a CLO test, and a Giemsa/silver stain. The frequencies of A2143G, A2142G, and the wild type of 23S rRNA and the presence of H. pylori were determined by Seeplex ClaR-H. pylori PCR (Seegene Inc., Seoul, Korea). Twenty-nine culture isolates were tested for susceptibility to clarithromycin by E-test (AB Biodisk, Solna, Sweden) or the CLSI (Clinical and Laboratory Standards Institute) disk diffusion test.
Results
From 176 H. pylori PCR-positive specimens, 23S rRNA gene mutations were detected in 38 isolates (21.6%), including 27 isolates of A2143G and 11 isolates of A2142G. Total mutation rates varied from 15.8% to 31.3% with the frequency of A2143G mutation alone varying from 8.5% to 25.0% among the five hospitals studied. There were 10 clarithromycin-resistant isolates found by susceptibility test and they were all positive for A2143G mutation. But, 3 of the 19 susceptible isolates were also positive for either A2143G or A2142G mutation.
Go to : 
References
2. Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, et al. Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med. 1991; 325:1127–31.
3. Suerbaum S and Michetti P. Helicobacter pylori infection. N Engl J Med. 2002; 347:1175–86.
4. Pounder RE and Ng D. The prevalence of Helicobacter pylori infection in different countries. Aliment Pharmacol Ther. 1995; 9:33–9.
5. Yim JY, Kim N, Choi SH, Kim YS, Cho KR, Kim SS, et al. Seroprevalence of Helicobacter pylori in South Korea. Helicobacter. 2007; 12:333–40.

6. Chung WC, Cho YS, Jeong JJ, Lee IS, Kim SW, Yang JM, et al. Eradication rate of Helicobacter pylori according to the diseases and therapeutic regimens, and reinfection rate after successful eradication in a tertiary clinic. Korean J Gastroenterol. 2003; 41:1–8.
7. Kim JJ, Reddy R, Lee M, Kim JG, EI-Zaatari FA, Osato MS, et al. Analysis of metronidazole, clarithromycin and tetracycline resistance of Helicobacter pylori isolates from Korea. J Antimicrob Chemother. 2001; 47:459–61.

8. Kim ES, Kang JO, Han D, Park PW, Park IK, Choi TY. Comparison of modified broth microdilution method, E test and disk diffusion method for antimicrobial susceptibility testing of Helicobacter pylori. Korean J Clin Pathol. 1998; 18:559–64.
9. Houben MH, van de Beek D, Hensen EF, Craen AJ, Rauws EA, Tytgat GN. A systematic review of Helicobacter pylori eradication therapy-the impact of antimicrobial resistance on eradication rates. Aliment Pharmacol Ther. 1999; 13:1047–55.

10. Grignon B, Tankovic J, Mégraud F, Glupczynski Y, Husson MO, Conroy MC, et al. Validation of diffusion methods for macrolide susceptibility testing of Helicobacter pylori. Microb Drug Resist. 2002; 8:61–6.
11. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; eighteenth informational supplement. M2-A9 and M7-A7, M100-S18. Wayne, PA; CLSI. 2008.
12. Kim JM, Kim JS, Jung HC, Kim N, Song IS. Antibiotic resistance of Helicobacter pylori isolated from Korean patients in 2003. Korean J Gastroenterol. 2004; 44:126–35.
13. Kim JM, Kim JS, Jung HC, Kim N, Kim YJ, Song IS. Distribution of antibiotic MICs for Helicobacter pylori strains over a 16-year period in patients from Seoul, South Korea. Antimicrob Agents Chemother. 2004; 44:4843–7.
14. Sung H, Chung HJ, Kim MN, Lee GH. Clinical usefulness of antimicrobial susceptibility test for Helicobacter pylori. Korean J Lab Med. 2006; 26:179–84.

15. Occhialini A, Urdaci M, Doucet-Populaire F, Bébéar CM, Lamouliatte H, Mégraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997; 41:2724–8.

16. Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, et al. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996; 40:477–80.

17. Taylor DE, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of two copies of a 23S rRNA gene from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997; 41:2621–8.

18. Alarcón T, Domingo D, Prieto N, López-Brea M. PCR using 3'-mismatched primers to detect A2142C mutation in 23S rRNA conferring resistance to clarithromycin in Helicobacter pylori clinical isolates. J Clin Microbiol. 2000; 38:923–5.
19. van Doorn LJ, Glupczynski Y, Kusters JG, Mégraud F, Midolo P, Maggi-Solca N, et al. Accurate prediction of macrolide resistance in Helicobacter pylori by a PCR line probe assay for detection of mutations in the 23S rRNA gene: multicenter validation study. Antimicrob Agents Chemother. 2001; 45:1500–4.
20. Stone GG, Shortridge D, Versalovic J, Beyer J, Flamm RK, Graham DY, et al. A PCR-oligopeptide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997; 41:712–4.
21. Mégraud F, Lehn N, Lind T, Bayerdörffer E, O'Morain C, Spiller R, et al. Antimicrobial susceptibility testing of Helicobacter pylori in a large multicenter trial: the MACH 2 study. Antimicrob Agents Chemother. 1999; 43:2747–52.
22. Umegaki N, Shimoyama T, Nishiya D, Suto T, Fukuda S, Munakata A. Clarithromycin-resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Japan. J Gastroenterol Hepatol. 2000; 15:906–9.
23. Maeda S, Yoshida H, Matsunaga H, Ogura K, Kawamata O, Shiratori Y, et al. Detection of clarithromycin-resistant Helicobacter pylori strains by a preferential homoduplex formation assay. J Clin Microbiol. 2000; 38:210–4.
24. Wang WH, Wong BCY, Mukhopadhyay AK, Berg DE, Cho CH, Lai KC, et al. High prevalence of Helicobacter pylori infection with dual resistance to metronidazole and clarithomycin in Hong Kong. Aliment Pharmacol Ther. 2000; 14:901–10.
25. Lee MA. Antimicrobial resistance of Helicobacter pylori. Korean J Clin Microbiol. 2001; 4:73–7.
Go to : 
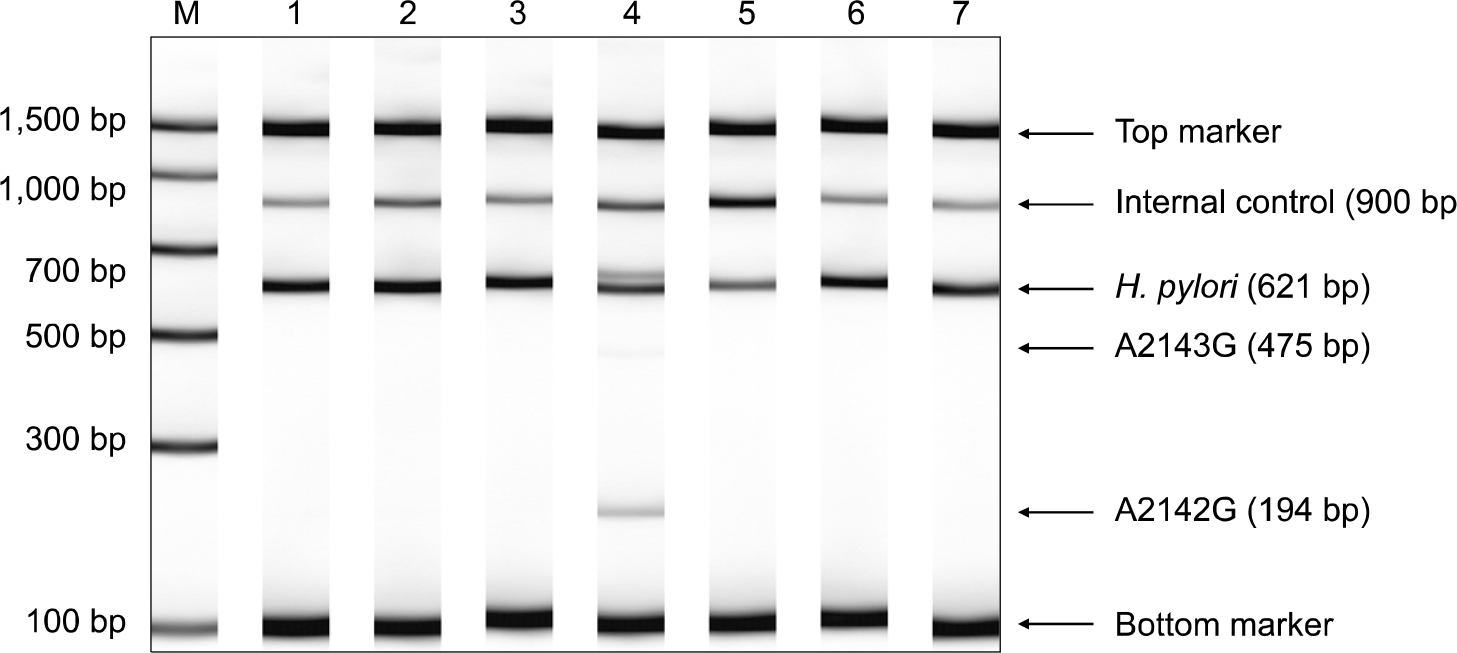 | Fig. 1.Detection of the point mutations at 2143 (A to G) and 2142 (A to G) in 23S rRNA gene by polymerase chain reaction products. Lane M, DNA ladder; lane 1–3 and 5–7, wild type; lane 4, mutant type of A2142G. |
Table 1.
Endoscopic diagnosis of patients at 5 university hospitals in Korea
| Hospital | Gastritis | Gastric ulcer | Gastric polyp/adenoma | Gastric cancer | Total |
|---|---|---|---|---|---|
| A | 18 | 8 | 0 | 10 | 36 |
| B | 76 | 13 | 0 | 4 | 93 |
| C | 31 | 3 | 0 | 0 | 34 |
| D | 77 | 20 | 0 | 3 | 100 |
| E | 69 | 13 | 6 | 2 | 90 |
| Total | 271 | 57 | 6 | 19 | 353 |
Table 2.
Comparison of Seeplex ClaR-H. pylori PCR and other methods
| Seeplex ClaR-H. pylori PCR | CLO or Giemsa/silver stain or culture | |
|---|---|---|
| Positive | Negative | |
| Positive | 163∗ | 13∗ |
| Negative | 31∗ | 146∗ |
| Total | 194∗ | 159∗ |
Table 3.
Allele frequencies of A2143G and A2142G at five university hospitals in Korea




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download