1. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007; 17:118–126. PMID:
17197185.

2. Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012; 13:271–282. PMID:
22411466.

3. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004; 101:2999–3004. PMID:
14973191.

4. Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007; 131:1109–1123. PMID:
18083101.

5. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007; 449:682–688. PMID:
17898713.

6. Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008; 451:147–152. PMID:
18185580.

7. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009; 139:871–890. PMID:
19945376.

8. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007; 7:415–428. PMID:
17508028.

9. Nieto MA, Cano A. The epithelial-mesenchymal transition under control: global programs to regulate epithelial plasticity. Semin Cancer Biol. 2012; 22:361–368. PMID:
22613485.

10. Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem. 2014; 60:197–205. PMID:
24132943.

11. Gwak JM, Kim HJ, Kim EJ, Chung YR, Yun S, Seo AN, et al. Micro RNA-9 is associated with epithelial-mesenchymal transition, breast cancer stem cell phenotype, and tumor progression in breast cancer. Breast Cancer Res Treat. 2014; 147:39–49. PMID:
25086633.
12. Chistiakov DA, Sobenin IA, Orekhov AN, Bobryshev YV. Human miR-221/222 in physiological and atherosclerotic vascular remodeling. Biomed Res Int. 2015; 2015:354517. PMID:
26221589.

13. Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012; 12:27–33. PMID:
22082479.

14. Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007; 282:23716–23724. PMID:
17569667.

15. Falkenberg N, Anastasov N, Rappl K, Braselmann H, Auer G, Walch A, et al. MiR-221/-222 differentiate prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer. 2013; 109:2714–2723. PMID:
24129242.

16. Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010; 102:706–721. PMID:
20388878.
17. Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011; 30:1082–1097. PMID:
21057537.

18. Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011; 4:ra41. PMID:
21673316.

19. Choi Y, Lee HJ, Jang MH, Gwak JM, Lee KS, Kim EJ, et al. Epithelial-mesenchymal transition increases during the progression of in situ to invasive basal-like breast cancer. Hum Pathol. 2013; 44:2581–2589. PMID:
24055090.

20. Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ, et al. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol. 2011; 22:1736–1747. PMID:
21709140.

21. Lee C, He H, Jiang Y, Di Y, Yang F, Li J, et al. Elevated expression of tumor miR-222 in pancreatic cancer is associated with Ki67 and poor prognosis. Med Oncol. 2013; 30:700. PMID:
24026657.

22. Xu K, Liang X, Shen K, Sun L, Cui D, Zhao Y, et al. MiR-222 modulates multidrug resistance in human colorectal carcinoma by down-regulating ADAM-17. Exp Cell Res. 2012; 318:2168–2177. PMID:
22677042.

23. Sun C, Li N, Zhou B, Yang Z, Ding D, Weng D, et al. miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27(kip1.). Oncol Lett. 2013; 6:507–512. PMID:
24137356.

24. Brisken C, O'Malley B. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2010; 2:a003178. PMID:
20739412.

25. Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, et al. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014; 19:7122–7137. PMID:
24886939.

26. Sarrió D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008; 68:989–997. PMID:
18281472.

27. Jeong H, Ryu YJ, An J, Lee Y, Kim A. Epithelial-mesenchymal transition in breast cancer correlates with high histological grade and triple-negative phenotype. Histopathology. 2012; 60:E87–E95. PMID:
22439911.

28. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011; 121:2750–2767. PMID:
21633166.

29. Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008; 283:29897–29903. PMID:
18708351.

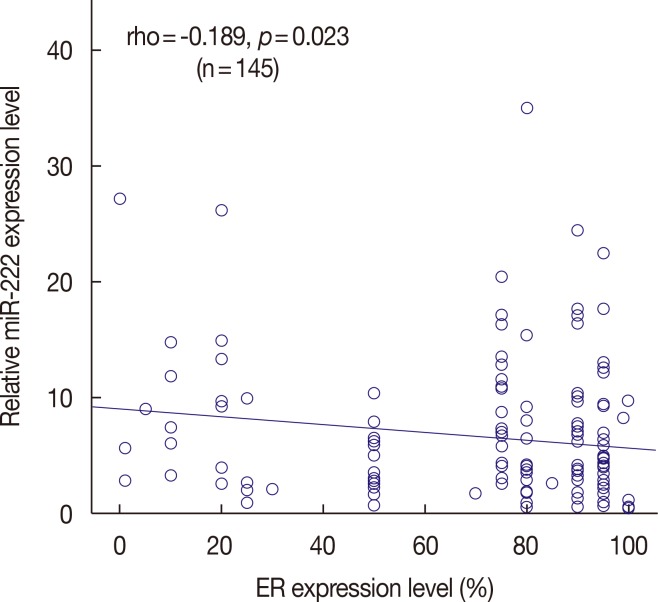
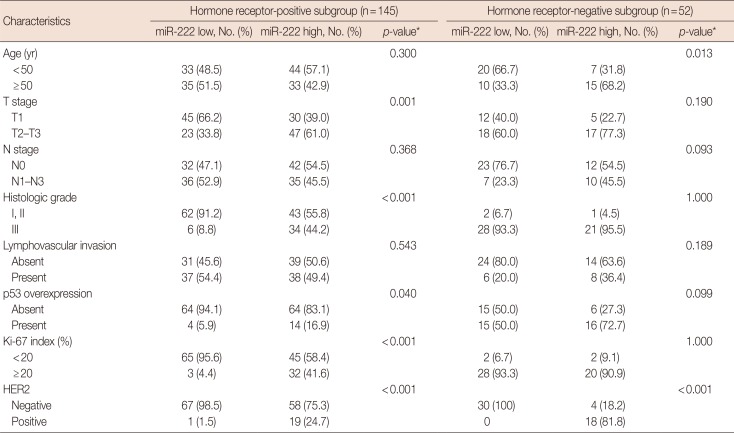
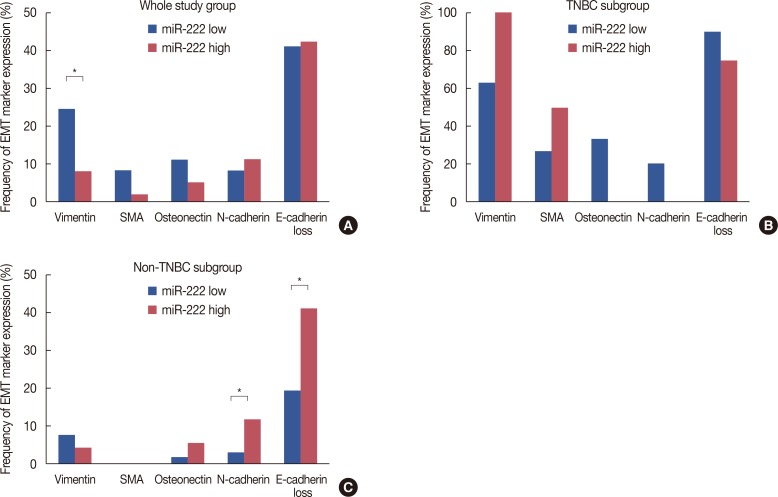
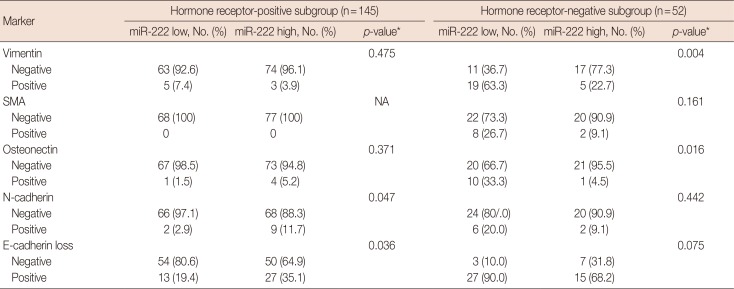
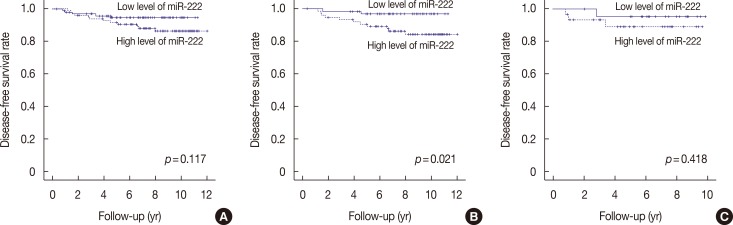
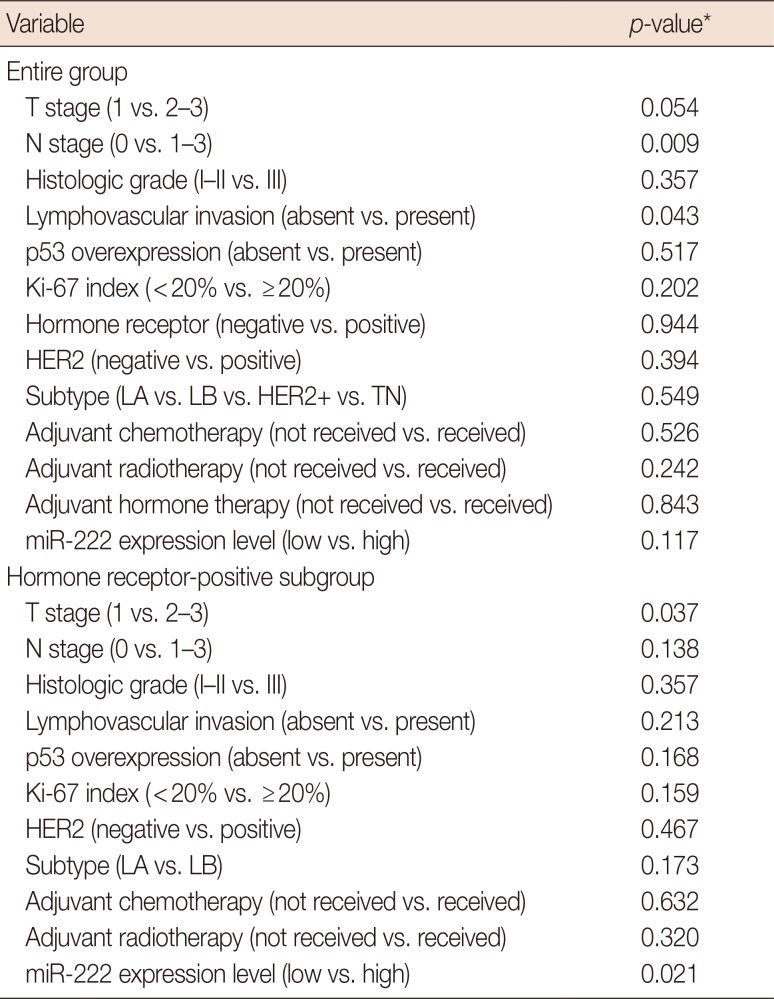
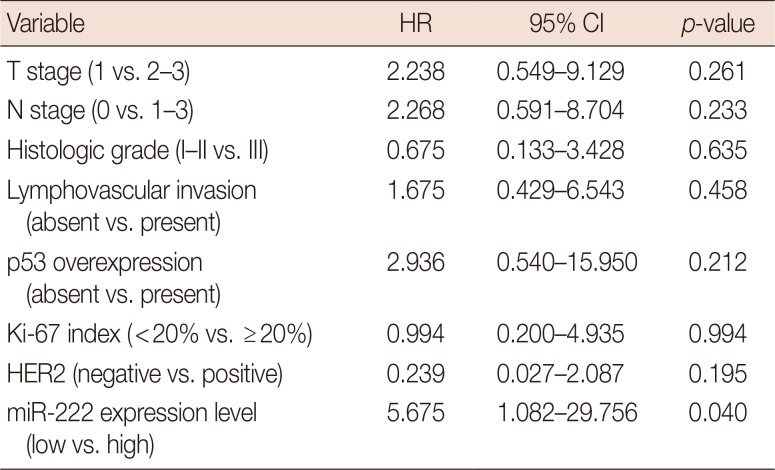




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download