Abstract
Activation of the mammalian target of rapamycin (mTOR) signaling pathway is an important mechanism of resistance to endocrine therapy in breast cancer. Everolimus, an mTOR inhibitor, has been shown to increase the efficacy of endocrine therapy and overcome resistance to endocrine therapies. Clinical studies have suggested that everolimus combined with endocrine therapy prolongs progression-free survival in hormone receptor-positive breast cancer patients. However, because breast cancer includes a group of highly heterogeneous tumors, patients may have different responses to everolimus. Therefore, finding biomarkers that can predict a patient's positive response or resistance to everolimus is critical. Numerous preclinical studies have shown that PIK3CA/PTEN mutations are predictive of sensitivity to everolimus; however, clinical trials have not confirmed the correlation between mutation status and clinical response. KRAS or BRAF mutations can bypass the phosphatidylinositol 3-kinase pathway; therefore, mutations in KRAS or BRAF may lead to resistance to mTOR inhibitors, and preclinical studies have shown that PIK3CA mutant cells which also contain KRAS mutations are resistant to everolimus. However, there are no clinical data in breast cancer patients to support this conclusion. Therefore, large-scale clinical studies are needed to identify biomarkers of efficacy and resistance to everolimus.
Breast cancer is the most common malignant tumor in women worldwide, thus it is a serious threat to women's health [12]. Most breast tumors express hormone receptors (HR), including estrogen receptor and progesterone receptor, and can benefit from endocrine therapy [3]. However, approximately 25% of HR-positive advanced breast cancers show primary or secondary resistance to endocrine therapy [4]. Previous studies have revealed that the phosphatidylinositol 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway is involved in the mechanisms of resistance to endocrine therapy [5], and mTOR inhibitors have been shown to have the potential to overcome resistance to endocrine therapy [56]. As a selective inhibitor of mTOR, everolimus in combination with endocrine therapy could prolong the progression-free survival (PFS) of patients with HR-positive human epidermal growth factor receptor 2 (HER2)-negative breast cancer [789]. However, biomarkers that can help select patients who will most benefit from everolimus or those that show resistance to everolimus are urgently needed [10]. This article briefly summarizes the clinical trials of everolimus and reviews the potential biomarkers of everolimus response in HR-positive breast cancer.
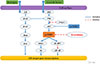
mTOR, which acts downstream of the PI3K/AKT pathway, is a serine/threonine protein kinase (Figure 1); as a result of its strategic position, mTOR is an important regulator of many cellular functions [1112]. mTOR has two major downstream messengers, ribosomal p70 S6 protein kinase 1 (S6K1) and 4E-binding protein (4E-BP1) [11]. Dysregulation of the PI3K/AKT signaling pathway is common in cancer, and this dysregulation can upregulate the mTOR pathway. Alteration of genes in the PIK3CA/AKT pathway is also a frequent occurrence in breast cancer, and the frequency of PI3K somatic mutations in breast cancers has been reported to be 20% to 45% [1314151617]. Furthermore, phosphatase and tensin homolog (PTEN) can inhibit the activity of the PI3K/AKT pathway, and PTEN gene loss has been reported in 15% of breast cancer patients [12]. Numerous studies have shown that tumors can become resistant to endocrine therapy through activation of this pathway. In addition, some preclinical and clinical studies showed that the addition of an mTOR inhibitor to conventional endocrine therapy could restore sensitivity to previously resistant tumor cells and improve disease treatment and the overall survival (OS) of patients with HR-positive breast cancers [1819].
Early-phase clinical trials suggested that everolimus may be an optional treatment for HR-positive metastatic breast cancers. A phase I study evaluated the pharmacokinetics and safety of everolimus plus letrozole in patients with metastatic breast cancer. In the study, seven out of 18 patients received combination therapy for more than 6 months. Prominent clinical toxicities were fatigue, stomatitis, diarrhea, anorexia, rash, and headache [20].
Additionally, a phase II study evaluated the safety and efficacy of fulvestrant combined with everolimus for patients with postmenopausal advanced breast cancer that was resistant to aromatase inhibitor. The median time to progression (TTP) in this study was 7.4 months, and the clinical benefit rate (CBR) was 49% [21]. In addition, 71% of the patients in this study received prior chemotherapy, 81% received prior tamoxifen therapy, and 26% received three or more types of endocrine therapy [21]. The most common adverse reactions were mucositis, weight loss, and rash. This study demonstrated that everolimus combined with fulvestrant is effective after aromatase inhibitor resistance in patients with heavily pretreated HR-positive breast cancer, and the toxicities were manageable [21].
The TAMRAD study is a randomized phase II study [9] on patients with HR-positive, HER2-negative metastatic breast cancer who were treated with prior aromatase inhibitor therapy. The purpose of the study was to evaluate the efficacy and safety of everolimus plus tamoxifen compared to tamoxifen alone. The results showed CBRs of 61% and 41% for the combination arm and the tamoxifen monotherapy arm, respectively, and this difference was statistically significant (p=0.04). The TTP was 4.5 months in patients treated with tamoxifen alone and 8.6 months in patients treated with everolimus plus tamoxifen. The prominent clinical toxicities reported in the combination arm were stomatitis, fatigue, rash, anorexia, and diarrhea. There was no difference in grade 3 or 4 adverse events between the two groups. Subgroup analysis of primary and secondary hormone resistance indicated that the median TTP was 14.8 months in patients with secondary resistance versus 5.4 months for patients with primary resistance. Similarly, patients with secondary resistance to aromatase inhibitors had a significantly higher CBR when treated with everolimus combined with tamoxifen (74%) than those treated with tamoxifen alone (48%). This study revealed that combination therapy with tamoxifen and everolimus increased the TTP, CBR, and OS when compared to tamoxifen monotherapy in patients with aromatase inhibitor-resistant postmenopausal metastatic breast cancer [9].
The promising results observed in the phase II studies of everolimus warranted further studies in patients with HR-positive, HER2-negative metastatic breast cancers, especially patients with de novo resistance to aromatase inhibitors. The BOLERO-2 study is a phase III study that compared everolimus combined with exemestane to placebo plus exemestane for patients with HR-positive HER2-negative breast cancer that is resistant to nonsteroidal aromatase inhibitor therapy. The results from the final analysis, after a median follow-up of 18 months, indicated that the primary end-point median PFS was 7.8 months in the everolimus plus exemestane arm versus 3.2 months in the placebo plus exemestane arm (p=0.0001). The CBR in the everolimus arm was significantly higher than that in the placebo arm (51.3% vs. 26.4%, p=0.0001) [7]. The median OS was 31.0 in patients treated with everolimus plus exemestane and 26.6 months in those treated with placebo plus exemestane; however, this difference was not statistically significant (p=0.1426) [8].
Tumors harboring mutations in genes encoding proteins involved in the PI3K/AKT/mTOR pathway may activate the PI3K enzyme. Therefore, such tumors are expected to be sensitive to everolimus and agents targeting this pathway. The results of several preclinical studies have suggested that genetic aberrations in the PI3K/AKT/mTOR pathway could predict the efficacy of mTOR inhibitors [2223]. However, data from clinical studies regarding the predictive capability of these genetic aberrations are contradictory. A retrospective study showed that patients with advanced breast cancer treated with inhibitors of the PI3K/AKT/mTOR pathway in combination with endocrine therapy, anti-HER2 therapy, or chemotherapy had a longer TTP compared to patients with wild-type tumors [24]. Baselga et al. [25] analyzed the relationship between the presence of mutations in exon 9 of PIK3CA and the efficacy of everolimus plus letrozole in a neoadjuvant trial. The results from the study indicated that the presence of these PIK3CA mutations provided an improved response to the combination of everolimus with letrozole [25]. However, in the TAMRAD trial, the researchers analyzed the relationship between mutations in the primary tumor tissue and the efficacy of everolimus, and they did not find any correlation between the presence of PI3K mutations and the response to everolimus [26]. Although they did find that everolimus was more effective in patients with low PI3K expression. In addition, patients with low levels of liver kinase B1 (LKB1), a known suppressor of mTOR, and high levels of phospho-4E binding protein, which is downstream of mTOR, received a greater benefit from everolimus treatment [26]. The BOLERO-2 study also failed to identify any specific gene mutations in the tumor tissue that were associated with a greater benefit from everolimus treatment [27]. Nevertheless, the investigators found that patients with no alteration or a single genetic alteration in PIK3CA/PTEN/CCND1 or FGFR1/2 received a greater PFS benefit from everolimus treatment [28]. In the BOLERO-2 study, mutation analysis of plasma cell-free DNA (cfDNA) suggested that there was no relationship between the PFS of patients treated with everolimus and the PIK3CA genotypes in cfDNA, which was consistent with previous tumor tissue DNA analysis. These results suggest that PIK3CA mutations, including H1047R, E545K, and E542K, cannot predict patient response to everolimus. In conclusion, the current evidence is insufficient to demonstrate that the PIK3CA genotype is an effective predictive biomarker for everolimus benefit [29].
Some PTEN gene variations, including germline and somatic mutations, can activate the PI3K enzyme. Preclinical models support the notion that cells with PTEN gene loss are more sensitive to PI3K/AKT inhibitors [3031]. However, the results from the TAMRAD trial showed that PTEN gene loss did not influence the response to everolimus [26]. In a similar analysis in the BOLERO-2 trial, PTEN gene status was not correlated with clinical outcome, which seems to confirm the results reported by the TAMRAD study [27].
In one study, a panel of breast cancer cell lines with HER2 amplification was sensitive to everolimus [32]. Another study showed that the combined presence of HER2 gene amplification along with PIK3CA mutation was highly predictive of sensitivity to an inhibitor of the PI3K/AKT/mTOR pathway (GDC-0941) [31]. Preclinical studies showed that the mTOR pathway is regulated by neurofibromin, a protein encoded by the NF1 gene, and several other studies demonstrated that cells containing mutations in the NF1 gene were highly sensitive to mTOR inhibitors, including everolimus and rapamycin [33]. A recent study explored the genomic alterations that confer extreme sensitivity to everolimus in 39 tumors of various types from patients who were treated with everolimus. The results showed that patients with mTOR, NF1, PIK3CA, TSC1, TSC2, and PIK3CG mutations could benefit from the mTOR inhibitor everolimus. Conversely, BAP1 and FGFR4 mutations were noted only in patients who did not receive a clinical benefit from everolimus. However, there were no breast cancer patients enrolled in this study [34]. In the BOLERO-2 study, researchers explored the correlation between ESR1 mutations, including Y537S and D538G, in cfDNA and sensitivity to exemestane and everolimus. Interestingly, these two activating mutations of the ESR1 gene appeared to have differential effects on sensitivity to everolimus. The results indicated that patients with the Y537S or D538G mutations had a poorer prognosis and shorter OS; however, the PFS of patients treated with everolimus was lower in those with the Y537S mutation than in those with wild-type ESR1, whereas the magnitudes of the PFS benefit of everolimus in patients with wild-type ESR1 and those with the D538G mutation were similar [35].
Preclinical studies in tumor cell lines indicated that high levels of phosphorylated AKT, glycogen synthase kinase 3 β, and tuberous sclerosis complex 2 are correlated with increased sensitivity to everolimus [36]. O'Reilly et al. [37] investigated potential predictive protein biomarkers for sensitivity to everolimus in human and animal studies, and univariate analysis showed that the levels of pS6, total S6, pS6/total S6, and phospho-AKT (pAKT) were significantly correlated with sensitivity to everolimus. Further analysis found that the combination of high pAKT and high p235-S6/total S6 levels could be a predictor of sensitivity to everolimus, whereas low pAKT and low p235-S6/total S6 levels could be a predictor of insensitivity to everolimus [37]. The TAMRAD trial found that everolimus was more effective in patients who had low PI3K expression, low LKB1 expression, and high phospho-4E binding protein expression; however, they did not find a relationship between the presence of PI3K, PTEN, and pAKT mutations and the efficacy of everolimus [26]. Baselga et al. [25] evaluated tumor core biopsies before treatment and on day 15 of treatment in patients treated with letrozole and either everolimus or placebo in a neoadjuvant trial of everolimus. The results indicated that patients treated with everolimus had a statistically significant decrease in Ki-67 and pS6 levels [25].
Clinical studies of PI3K/AKT/mTOR pathway inhibitors indicate that a larger number of patients develop de novo resistance to this class of anticancer agents. Therefore, understanding the biological basis of this de novo resistance and identifying biomarkers of resistance to this therapy are very important. KRAS or BRAF mutant proteins can bypass the PI3K/AKT/mTOR pathway. Therefore, patients with KRAS or BRAF mutations may show resistance to PI3K/AKT/mTOR inhibitors. Indeed, preclinical studies suggest that cells containing KRAS mutations are insensitive to everolimus [38]. Interestingly, a study of an non-small cell lung cancer (NSCLC) cell line suggested that de novo resistance to everolimus mediated by KRAS mutation is mitigated by a coexisting LKB1-deficiency as well as p53 loss. Importantly, these data suggest that LKB1/KRAS-mutant NSCLCs might be sensitive to mTOR-targeted therapies. Mahoney et al. [39] investigated the correlations between KRAS or BRAF mutation status and the efficacy of a PIK3CA inhibitor in a phase I clinical trial. The results indicated that colorectal cancer patients with PIK3CA or KRAS mutations did not respond to therapy, whereas patients with PIK3CA mutant ovarian cancer that also carried a KRAS or BRAF mutation did respond to PIK3CA inhibitor therapy. This study supported the hypothesis that the effects of BRAF and KRAS mutations may differ among tumors. However, there is no evidence for a relationship between KRAS or BRAF mutation status and the efficacy of everolimus in breast cancer.
Acquired resistance to everolimus may also be mediated by genetic alterations generated under the selective pressure of this agent. Although no such genetic alterations were reported in human trials, Zunder et al. [40] performed a preclinical study in Saccharomyces cerevisiae and identified a mutational hotspot in the Ile800 area of the PIK3CA gene, which confers a 5- to 10-fold decrease in potency for a large panel of PI3K and mTOR inhibitors. Unlike tyrosine kinase inhibitors, these resistance mutations do not reside in the classic gatekeeper residues [40], and cfDNA analyses in the BOLERO-2 study showed that the ESR1 mutation Y537S may be a resistance biomarker of sensitivity to everolimus [35].
In summary, activation of the mTOR signaling pathway is an important mechanism of endocrine therapy resistance in breast cancer. As an mTOR inhibitor, everolimus has been shown to increase the efficacy of endocrine therapies and may overcome drug resistance in HR-positive metastatic breast cancer. Mutations in the PIK3CA/AKT/mTOR pathway and PTEN gene loss are frequently observed in breast cancer patients, and numerous preclinical studies have demonstrated that mutations in PIK3CA could be predictive biomarkers of everolimus efficacy [2223]. However, the clinical trials have not supported this. None of the clinical studies have found any association between PIK3CA mutation status and clinical response to everolimus. The results of cell line studies assessing the predictive value of PTEN gene loss for everolimus sensitivity have not been consistent, and the clinical studies also did not find any correlation between PTEN gene loss and everolimus efficacy. However, this may be due to the heterogeneity of the tumors and the small number of patients included in the studies. It is also possible that the discordance mutational status between primary tumors and metastatic sites influenced the results. Circulating tumor DNA could be a promising biomarker given its potential to overcome the complications associated with tumor heterogeneity. Preclinical studies indicate that PIK3CA mutant cells carrying KRAS mutations are resistant to everolimus. However, there are no clinical data in breast cancer patients to support this finding. Large-scale clinical studies are also needed to identify biomarkers for sensitivity and resistance to everolimus.
Figures and Tables
Figure 1
The PI3K/AKT/mTOR signaling pathway, showing cascading pathway activation and regulatory feedback loops.
PI3K=phosphatidylinositol 3-kinase; mTOR=mammalian target of rapamycin; ER=estrogen receptor; IRS=insulin-receptor substrate; PTEN=phosphatase and tensin homolog; Erk=extracellular signal-regulated kinase; 4E-BPI=4E-binding protein 1; S6K1=ribosomal protein S6 kinase.
Notes
References
1. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016; 66:115–132.

2. Torre LA, Sauer AM, Chen MS Jr, Kagawa-Singer M, Jemal A, Siegel RL. Cancer statistics for Asian Americans, Native Hawaiians, and Pacific Islanders, 2016: converging incidence in males and females. CA Cancer J Clin. 2016; 66:182–202.

3. Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet. 2013; 45:1446–1451.

4. Ma CX, Reinert T, Chmielewska I, Ellis MJ. Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer. 2015; 15:261–275.

5. Paplomata E, O'Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014; 6:154–166.

6. Margariti N, Fox SB, Bottini A, Generali D. “Overcoming breast cancer drug resistance with mTOR inhibitors”. Could it be a myth or a real possibility in the short-term future? Breast Cancer Res Treat. 2011; 128:599–606.

7. Yardley DA, Noguchi S, Pritchard KI, Burris HA 3rd, Baselga J, Gnant M, et al. Everolimus plus exemestane in postmenopausal patients with HR(+) breast cancer: BOLERO-2 final progression-free survival analysis. Adv Ther. 2013; 30:870–884.

8. Piccart M, Hortobagyi GN, Campone M, Pritchard KI, Lebrun F, Ito Y, et al. Everolimus plus exemestane for hormone-receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: overall survival results from BOLERO-2†. Ann Oncol. 2014; 25:2357–2362.

9. Bachelot T, Bourgier C, Cropet C, Ray-Coquard I, Ferrero JM, Freyer G, et al. Randomized phase II trial of everolimus in combination with tamoxifen in patients with hormone receptor-positive, human epidermal growth factor receptor 2-negative metastatic breast cancer with prior exposure to aromatase inhibitors: a GINECO study. J Clin Oncol. 2012; 30:2718–2724.

10. Wander SA, Hennessy BT, Slingerland JM. Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy. J Clin Invest. 2011; 121:1231–1241.

11. Villarreal-Garza C, Cortes J, Andre F, Verma S. mTOR inhibitors in the management of hormone receptor-positive breast cancer: the latest evidence and future directions. Ann Oncol. 2012; 23:2526–2535.

12. Dhillon S. Everolimus in combination with exemestane: a review of its use in the treatment of patients with postmenopausal hormone receptor-positive, HER2-negative advanced breast cancer. Drugs. 2013; 73:475–485.

13. Ahmad F, Badwe A, Verma G, Bhatia S, Das BR. Molecular evaluation of PIK3CA gene mutation in breast cancer: determination of frequency, distribution pattern and its association with clinicopathological findings in Indian patients. Med Oncol. 2016; 33:74.

14. Kalinsky K, Jacks LM, Heguy A, Patil S, Drobnjak M, Bhanot UK, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009; 15:5049–5059.

15. Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012; 14:R28.
16. Barbareschi M, Buttitta F, Felicioni L, Cotrupi S, Barassi F, Del Grammastro M, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007; 13:6064–6069.

17. Pérez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjöld B, Rutqvist LE, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007; 13:3577–3584.

18. Zagouri F, Sergentanis TN, Chrysikos D, Filipits M, Bartsch R. mTOR inhibitors in breast cancer: a systematic review. Gynecol Oncol. 2012; 127:662–672.

19. Shtivelband MI. Everolimus in hormone receptor-positive advanced breast cancer: targeting receptor-based mechanisms of resistance. Breast. 2013; 22:405–410.

20. Awada A, Cardoso F, Fontaine C, Dirix L, De Grève J, Sotiriou C, et al. The oral mTOR inhibitor RAD001 (everolimus) in combination with letrozole in patients with advanced breast cancer: results of a phase I study with pharmacokinetics. Eur J Cancer. 2008; 44:84–91.

21. Massarweh S, Romond E, Black EP, Van Meter E, Shelton B, Kadamyan-Melkumian V, et al. A phase II study of combined fulvestrant and everolimus in patients with metastatic estrogen receptor (ER)-positive breast cancer after aromatase inhibitor (AI) failure. Breast Cancer Res Treat. 2014; 143:325–332.

22. Serra V, Markman B, Scaltriti M, Eichhorn PJ, Valero V, Guzman M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008; 68:8022–8030.

23. Gonzalez-Angulo AM, Blumenschein GR Jr. Defining biomarkers to predict sensitivity to PI3K/Akt/mTOR pathway inhibitors in breast cancer. Cancer Treat Rev. 2013; 39:313–320.

24. Janku F, Wheler JJ, Westin SN, Moulder SL, Naing A, Tsimberidou AM, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J Clin Oncol. 2012; 30:777–782.

25. Baselga J, Semiglazov V, van Dam P, Manikhas A, Bellet M, Mayordomo J, et al. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J Clin Oncol. 2009; 27:2630–2637.

26. Treilleux I, Arnedos M, Cropet C, Wang Q, Ferrero JM, Abadie-Lacourtoisie S, et al. Translational studies within the TAMRAD randomized GINECO trial: evidence for mTORC1 activation marker as a predictive factor for everolimus efficacy in advanced breast cancer. Ann Oncol. 2015; 26:120–125.

27. Hortobagyi GN, Chen D, Piccart M, Rugo HS, Burris HA 3rd, Pritchard KI, et al. Correlative analysis of genetic alterations and everolimus benefit in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: results from BOLERO-2. J Clin Oncol. 2016; 34:419–426.

28. Helwick C. Search for biomarkers of mTOR inhibitor benefit in breast cancer fails to pan out. ASCO Post;Accessed October 17th, 2017. http://www.ascopost.com/issues/july-10-2013/search-for-biomarkers-of-mtor-inhibitor-benefit-inbreast-cancer-fails-to-pan-out.aspx.
29. Moynahan ME, Chen D, He W, Sung P, Samoila A, You D, et al. Correlation between PIK3CA mutations in cell-free DNA and everolimus efficacy in HR(+), HER2(−) advanced breast cancer: results from BOLERO-2. Br J Cancer. 2017; 116:726–730.

30. Hong DS, Bowles DW, Falchook GS, Messersmith WA, George GC, O'Bryant CL, et al. A multicenter phase I trial of PX-866, an oral irreversible phosphatidylinositol 3-kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2012; 18:4173–4182.

31. O'Brien C, Wallin JJ, Sampath D, GuhaThakurta D, Savage H, Punnoose EA, et al. Predictive biomarkers of sensitivity to the phosphatidylinositol 3' kinase inhibitor GDC-0941 in breast cancer preclinical models. Clin Cancer Res. 2010; 16:3670–3683.
32. Hurvitz SA, Kalous O, Conklin D, Desai AJ, Dering J, Anderson L, et al. In vitro activity of the mTOR inhibitor everolimus, in a large panel of breast cancer cell lines and analysis for predictors of response. Breast Cancer Res Treat. 2015; 149:669–680.

33. Johannessen CM, Reczek EE, James MF, Brems H, Legius E, Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005; 102:8573–8578.

34. Lim SM, Park HS, Kim S, Kim S, Ali SM, Greenbowe JR, et al. Next-generation sequencing reveals somatic mutations that confer exceptional response to everolimus. Oncotarget. 2016; 7:10547–10556.

35. Chandarlapaty S, Sung P, Chen D, He W, Samoila A, You D, et al. cfDNA analysis from BOLERO-2 plasma samples identifies a high rate of ESR1 mutations: exploratory analysis for prognostic and predictive correlation of mutations reveals different efficacy outcomes of endocrine therapy-based regimens. In : 38th Annual CTRC-AACR San Antonio Breast Cancer Symposium; 2015. Abstract #S2-07.
36. Breuleux M, Klopfenstein M, Stephan C, Doughty CA, Barys L, Maira SM, et al. Increased AKT S473 phosphorylation after mTORC1 inhibition is rictor dependent and does not predict tumor cell response to PI3K/mTOR inhibition. Mol Cancer Ther. 2009; 8:742–753.

37. O'Reilly T, McSheehy PM. Biomarker development for the clinical activity of the mTOR inhibitor everolimus (RAD001): processes, limitations, and further proposals. Transl Oncol. 2010; 3:65–79.
38. Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, Macarulla T, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010; 120:2858–2866.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download