Abstract
Germline mutations in the BRCA1 and BRCA2 genes are strong genetic factors for predispositions to breast, ovarian, and other related cancers. This report describes a family with a history of breast and ovarian cancers that harbored a novel BRCA1 germline mutation. A single nucleotide deletion in intron 20, namely c.5332+4delA, was detected in a 43-year-old patient with breast cancer. This mutation led to the skipping of exon 20, which in turn resulted in the production of a truncated BRCA1 protein that was 1773 amino acids in length. The mother of the proband had died due to ovarian cancer and had harbored the same germline mutation. Ectopically expressed mutant BRCA1 protein interacted with the BARD1 protein, but showed a reduced transcriptional function, as demonstrated by the expression of cyclin B1. This novel germline mutation in the BRCA1 gene caused familial breast and ovarian cancers.
Hereditary breast cancers occur in about 5% to 10% of patients with breast cancers [12]. Germline mutations occurring in the BRCA1 and BRCA2 genes have been identified as the causal mutations for hereditary breast, ovarian, and other related cancers [34]. Carriers of BRCA1 mutations are estimated to have lifetime risks of 87% and 44% for breast and ovarian cancers, respectively [56]. The most pathogenic mutations in the BRCA1 and BRCA2 genes are the nonsense mutations and the small insertions and deletions, which lead to the translation of truncated proteins [78]. Furthermore, splicing analysis of the BRCA1 and BRCA2 variants has revealed that several mutations at the exon-intron boundaries disrupt the accuracy of splicing of the corresponding mRNAs, resulting in altered splicing events [910]. Point mutations and nonsense mutations in the BRCA1 gene that cause the skipping of one or more exons have been reported in patients with cancers [11]. Here, we report a novel germline mutation in the BRCA1 gene that results in exon 20 skipping in a family with a history of breast and ovarian cancers.
A 43-year-old woman with breast cancer visited the National Cancer Center in Korea for obtaining treatment, and she was referred to the genetic counseling clinic, due to her family history of breast and ovarian cancers. Her pedigree was assessed, and blood samples were collected from 17 family members. The proband had been diagnosed with breast cancer at 43 years of age, and her mother had died of ovarian cancer at an age of 66 years. Among the maternal relatives of the proband, two cousins had been diagnosed with breast cancers, but further information was not available (Figure 1). The proband underwent nipple-sparing mastectomies of both breasts, and immunohistochemistry revealed that her tumor tissue was of a triple negative breast cancer subtype, i.e., negative for estrogen and progesterone receptors and for human epidermal growth factor receptor 2. Despite strong recommendations by her doctors, she refused an oophorectomy, but she did undergo regular gynecological examinations, including trans-vaginal ultrasounds.
The genetic testing of the BRCA1 and BRCA2 genes in the proband was performed by using a direct sequencing method. Genetic testing of the proband showed a single nucleotide deletion in the BRCA1 intron 20, namely c.5332+4delA (Figure 2A). To test the mRNA sequence of BRCA1, total RNA was extracted from proband's blood, and the cDNAs corresponding to BRCA1 exons 19, 20, and 21 were sequenced. RNA sequencing showed that the abovementioned deletion had resulted in exon 20 skipping and the introduction of a premature stop codon (Figure 2B). The mother of the proband had died of ovarian cancer and had harbored the same mutation. Among 17 other family members, two uncles and two cousins too harbored the same mutation.
We examined whether the mutation had a functional effect on the BRCA1 protein. Mutant (exon 20 deletion) and wild-type (WT) BRCA1 were overexpressed in a breast cancer cell line, MDA-MB-436 cells, and the resultant protein expression levels were estimated by Western blotting. Exogenous expression of these proteins in MDA-MB-436 cells revealed that the levels of the mutant protein were much lower than those of WT BRCA1. Following treatment of the transfected cells with MG132 to block potential proteasomal degradation, the levels of mutant and WT proteins in these cells were comparable (Figure 3A). To assess whether mutant BRCA1 could interact with BARD1, BRCA1, and BARD1 proteins were co-immunoprecipitated as described previously [12]. Both the WT and mutant proteins immunoprecipitated with BARD1 (Figure 3B), in proportion to their level of expression. The levels of cyclin B1 mRNA in these cells were measured by real-time reverse transcription polymerase chain reaction, following the exogenous expression of WT or mutant BRCA1. WT BRCA1 enhanced the expression levels of cyclin B1 mRNA; however, in cells that overexpressed mutant BRCA1 with or without WT, the levels of cyclin B1 mRNA remained unchanged (Figure 3C).
Germline mutations of the BRCA1 and BRCA2 genes, which result in truncated proteins, have been identified as the pathogenic mutations that cause the hereditary cancers. Carriers of the BRCA1 and BRCA2 mutations should be monitored carefully, due to the greatly increased risks of the incidence of cancers in these individuals.
Here, we report a patient who harbored a single nucleotide deletion in intron 20 of the BRCA1 gene. The mother of this patient had ovarian cancer and harbored the same mutation. Among 17 other family members, two uncles and two cousins too harbored the mutation.
A single nucleotide deletion at the 5' end of intron 20 caused an altered splicing event, which resulted in the production of an altered sequence beginning at the amino acid at position 1760 in the sequence; this, in turn, resulted in the translation of a truncated protein with 1773 amino acids. A truncated BRCA1 protein with a short C-terminal sequence may display an abnormal protein lacking the BRCA1 C-terminal (BRCT) domain, which has been reported to be essential for the tumor suppressor function and protein interaction of BRCA1 [13]. Splicing variants in exons 19–21 of the BRCA1 gene that cause exon skipping or intron retention due to aberrant splicing have been reported in patients with breast cancers [1014].
Functional analyses of the mutant BRCA1 protein (exon 20 deletion) revealed that the protein levels of this mutant were lesser than those of the WT BRCA1. Furthermore, the results of inhibiting proteasomal degradation following the exogenous expression of these proteins suggested that the mutant protein was more susceptible to proteasomal degradation. Results of immunoprecipitation studies showed that both the mutant and WT BRCA1 could interact with BARD1. However, the mutation impaired the transcriptional activity of BRCA1. To assess the effect of the c.5332+4delA mutation on the transcriptional function of BRCA1, the levels of cyclin B1 mRNA were compared between cells that exogenously expressed the WT and mutant BRCA1 proteins. Consistent with previous reports [15], the WT BRCA1 altered the expression of cyclin B1 mRNA. However, the mutant BRCA1 could not alter the levels of cyclin B1 mRNA (Figure 3C). Interestingly, cells expressing both WT and mutant BRCA1 showed normal levels of cyclin B1 mRNA, suggesting that the mutant protein has a dominant negative effect.
In conclusion, this report describes a novel germline mutation of BRCA1 that causes exon 20 skipping in a Korean family with a history of breast cancer. These data suggest that germline mutations occurring in splice site consensus sequences should be investigated carefully in families with histories of cancers.
Figures and Tables
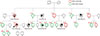 | Figure 1Pedigree of the patient's family, showing multiple individuals with breast and ovarian cancer. The proband (arrow) and her family members harboring the mutation (c.5332+4delA) in BRCA1 gene are marked in red. Male is represented by a square and female by a circle. Numbers in parentheses indicate age at cancer diagnosis. |
 | Figure 2Mutation analysis of exon 20 in BRCA1 gene. (A) Sequence analysis of the mutant BRCA1 gene shows a deletion in c.5332+4delA (genomic DNA) and exon 20 skipping (RNA). Exon 20 sequences are colored in yellow. (B) The schematic view shows the variant localization and the mutated mRNA skipping 55 bp of exon 20. |
 | Figure 3Functional analyses of mutant BRCA1 protein. (A) Mutant BRCA1 protein (exon 20 deletion) showed reduced protein level. BRCA1-deficient MDA-MB-436 cells were transfected with a vector expressing HA-tagged BRCA1 wild-type (WT) or exon20 skipping mutant or untagged mutant BRCA1 (exon 20 deletion) mRNA. A point mutation of BRCA1 that causes an amino acid substitution of leucine to phenylalanine (L52F) was used as a missense mutant control. After 24 hours, the cells were treated with DMSO (control) or MG132 (1 µM). The arrows indicate BRCA1 protein and the asterisk indicates a nonspecific band. (B) WT or mutant BRCA1 is immunoprecipitated with anti-BARD1 antibody. Immunoprecipitated WT and mutant BRCA1 were detected by Western blotting with an anti-BRCA1 antibody (boxed lane). (C) Expression level of cyclin B1 was compared among cells transfected with WT or mutant BRCA1. Relative expression of cyclin B1 was measured by real-time reverse transcription polymerase chain reaction compared to glyceraldehyde 3-phosphate dehydrogenase after exogenous expression of WT or mutant BRCA1.DMSO=dimethyl sulfoxide; Ex20del=exon 20 deletion; BARD1=BRCA1 associated RING domain1; GFP=green fluoresence protein (control DNA); IP=immunoprecipitation; IB=immunoblotting.
*Nonspecific band.
|
Notes
References
2. Lux MP, Fasching PA, Beckmann MW. Hereditary breast and ovarian cancer: review and future perspectives. J Mol Med (Berl). 2006; 84:16–28.

3. Hall MJ, Reid JE, Burbidge LA, Pruss D, Deffenbaugh AM, Frye C, et al. BRCA1 and BRCA2 mutations in women of different ethnicities undergoing testing for hereditary breast-ovarian cancer. Cancer. 2009; 115:2222–2233.

5. Couch FJ, DeShano ML, Blackwood MA, Calzone K, Stopfer J, Campeau L, et al. BRCA1 mutations in women attending clinics that evaluate the risk of breast cancer. N Engl J Med. 1997; 336:1409–1415.

6. Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE. Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet. 1994; 343:692–695.
7. Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007; 25:1329–1333.
8. Langston AA, Malone KE, Thompson JD, Daling JR, Ostrander EA. BRCA1 mutations in a population-based sample of young women with breast cancer. N Engl J Med. 1996; 334:137–142.

9. Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002; 3:285–298.

10. Houdayer C, Caux-Moncoutier V, Krieger S, Barrois M, Bonnet F, Bourdon V, et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat. 2012; 33:1228–1238.

11. Liu HX, Cartegni L, Zhang MQ, Krainer AR. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat Genet. 2001; 27:55–58.

12. Chang S, Biswas K, Martin BK, Stauffer S, Sharan SK. Expression of human BRCA1 variants in mouse ES cells allows functional analysis of BRCA1 mutations. J Clin Invest. 2009; 119:3160–3171.

13. Coquelle N, Green R, Glover JN. Impact of BRCA1 BRCT domain missense substitutions on phosphopeptide recognition. Biochemistry. 2011; 50:4579–4589.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download