Abstract
Purpose
Metastasis and local recurrence are the primary causes of treatment failure and patient death in breast cancer. The aim of this study was to validate a metastasis- and local recurrenceassociated biomarker for prognostic evaluation and planning treatment strategies.
Methods
Formalin-fixed, paraffin-embedded tissues from a cohort of 312 patients (all stage II and III) were used. The prevalence of CD49f+ cells in the patients' tumors was analyzed and correlated with clinical characteristics to determine its prognostic and clinical implications.
Results
CD49f+ tumor cells were found in a minority of tumors, with 62.8% of the samples showing not a single cell of this subtype. In the clinical characteristics analysis, which were performed with t-tests, CD49f+ tumors were not associated with age, tumor size, World Health Organization grade, nodal status, human epidermal growth factor receptor 2 status, progesterone receptor status, or estrogen receptor status, although they were significantly associated with disease recurrence (distant metastasis or/and local recurrence). Univariate survival analysis using the Kaplan-Meier method showed that CD49f+ tumors were associated with markedly decreased disease-free survival (DFS); the same result was found using multivariate Cox analysis, even when only chemotherapy-treated patients were analyzed.
Conclusion
Our results indicated that breast tumors with CD49f+ cancer cells are associated with an increased risk for disease recurrence after initial surgery with poor clinical outcomes (decreased DFS). Therefore, as it requires testing for only one additional protein, adding CD49f testing to conventional surgical pathology is a strategy that has great potential for prognostic and treatment-guidance purposes.
Metastasis is a landmark event for the breast cancer patients, which transforms a local neoplasm into a systemic, metastatic, and life-threatening disease. According to a report by the American Cancer Society, the 5-year relative survival rate for women with breast cancer decreases from 99% when diagnosed with only a localized tumor to 23% when diagnosed with distant metastasis [1]. Identification of metastasis protein biomarkers at an early stage can help to identify patients at high risk for metastasis, enhancing personalized therapy and increasing quality of life for tumor survivors. Previously, we identified aldehyde dehydrogenase (ALDH) 1A3, ALDH+/CD44+, and epithelial cell adhesion molecule (EpCAM)–/CD49f+ as prognostic biomarkers for breast cancer [23]. Although these markers are helpful for prognosis and treatment purposes, quality controls for dual immunohistochemistry (IHC) do not always meet clinical requirements.
Since metastasis and local recurrence are crucially important during breast cancer treatment, different testing techniques are used for prognosis and treatment purposes, including Oncotype DX® (based on the expression of 21 genes), MammaPrint (based on the expression of 70 genes), and the molecular subtyping system (based on progesterone receptor [PR]/estrogen receptor [ER]/human epidermal growth factor receptor 2 [HER2] expression), among others [45678]. All of the above systems detect abnormal gene expression, and most target treatments are based on blocking specific abnormally activated genes or signal pathways, such as trastuzumab for HER2-positive breast cancers. However, there are always compensatory signaling pathways on standby, and once a certain signal is blocked, alternative pathways will become activated under selection pressure, ultimately leading to treatment resistance [910]. Therefore, rather than screening for driver genes in tumors, we are more interested in searching for the cellular origins of metastasis and local recurrence.
Different models have been proposed to illustrate the dynamic process of how a localized tumor invades adjacent and distant tissues. One of the most well-known theories is the breast cancer stem cell (BCSC) theory. Intratumoral heterogeneity in breast cancer is well documented and widely accepted, and many researchers believe that the apparently heterogeneous populations of tumor cells are not in fact disorganized, but instead represent a well-organized hierarchy. Because BCSCs are normally dormant within the tumor and show resistance to chemotherapy, radiation therapy, and cytotoxins, identifying biomarkers for BCSCs and breast epithelial stem cells will be useful resources for studying local recurrence and metastasis in patients with poor clinical outcomes [11].
Because multiple markers are included in panels for identifying BCSCs, it can be difficult to apply such methods to routine surgical biopsy workup. Previously, Abraham et al. [11] used dual CD44+/CD24– IHC staining to label BCSCs and found that the prevalence of CD44+/CD24– tumor cells in breast cancer is correlated with distant metastasis. However, such techniques are not applicable for routine usage because of the complicated staining procedure and scoring criteria, as well as quality control issues.
Recently, Lim et al. [12] reported that the EpCAMlow/CD49f+ subset of cells shows mammary regeneration capacity based on transplantation studies in NOD-SCID mice. In particular, they showed that while the EpCAMlow/CD49f+ subpopulation was enriched for mammary stem cell (MaSC) (bipotent) progenitors, the EpCAM+/CD49f+ subpopulation was enriched for luminal progenitors [12]. Therefore, CD49f+ cells share certain phenotypic similarities with adult tissue stem cells/progenitors. We propose that by simply identifying CD49f+ cells in breast cancer tissue samples, we can identify high-risk metastasis patients at very early stages of diagnosis, which will positively contribute to personalized therapy and increase quality of life for breast cancer survivors.
We studied 312 breast tumor tissues from a cohort of 456 patients diagnosed with breast cancer who had the tumor removed at West China Hospital between 2006 and 2009. Disease-free survival (DFS) and overall survival (OS) were defined as the time between the initial surgery and local or distant metastatic relapse, and between surgery and death, respectively. We prepared tissue microarray (TMA) cores (1.5-mm diameter) from formalin-fixed paraffin-embedded samples. Two cores from every individual tumor were tested on the arrays. IHC staining was performed on all of the TMA slides and the results were interpreted by two pathologists using a blinded method. In cases of disagreement, they discussed it until they reached an agreement. Then, the mean score of the two cores was taken as the final score. Approval for this study was granted by the Ethics Committee of West China Hospital (IRB number: 2013-191).
The procedure for IHC staining was performed as previously reported [2]. Antibody detection of CD49f (TA506627, 1:150; OriGene, Rockville, USA) was performed using an automated staining machine according to the manufacturer's instructions. CD49f antibodies were detected with diaminobenzidine (DAB).
Hematoxylin and eosin stained slides and IHC slides were assessed by pathologists at West China Hospital. The pathologists conducted standard pathological tumor assessments, and the identities of the patients were unknown to the pathologists in all subsequent studies. The expression levels of epidermal growth factor receptor, cytokeratin 5/6, ER, PR, and HER2 were obtained from the pathology reports. HER2 staining was analyzed according to the American Society of Clinical Oncology guidelines. When the expression percentage of ER or PR was above 1%, we defined the case as “positive” [13].
To accurately qualify the number of target tumor cells in this tissue micro-array sample, the entire area of each tissue was counted. The scoring rules were as follows: 0, 0% positive tumor cells; 1, 1% to 10% positive cells; 2, 11% to 50% positive cells; 3, 51% to 75% positive cells; and 4, 76% to 100% positive cells. CD49f staining was detected mainly in the cytoplasm, and scoring was performed as described for CD24 [14].
Associations between the presence of CD49f+ and clinical variables as well as breast cancer subgroups were assessed using t-tests. The Kaplan-Meier method was used to estimate DFS and OS, and the log-rank test was used to compare survival between two strata. All tests were two-sided, and p<0.05 was considered as statistically significant. Multivariate survival analysis was performed using the Cox proportional hazard model. All statistical analyses were performed using the SPSS version 16.0 software program (SPSS Inc., Chicago, USA).
From the total group of 456 patients, 53 samples lacked follow-up information, 23 did not have primary tumor samples, and 61 patients were in clinical stage I and IV, yielding 319 samples that were processed for IHC staining. Finally, 312 samples passed the TMA slide quality assessment for IHC staining (Figure 1). All qualified patients (n=312) were women, ranging in age from 27 to 77 years (49.25±9.85 years). All patients (312/312, 100%) were diagnosed with invasive ductal carcinoma at clinical stage II or III. We performed a Kaplan-Meier analysis and used the log-rank test to determine the effect of clinicopathological characteristics on prognosis. As expected, World Health Organization (WHO) grade (OS, p<0.001), tumor size (OS, p<0.001), node status (OS, p<0.010), clinical stage (DFS, p<0.001; OS, p<0.001), PR status (DFS, p=0.030; OS, p=0.003), and presence of CD49f (DFS, p=0.024) were significantly correlated with a poor clinical outcome (Table 1).
We performed IHC staining on the TMA tissues, and CD49f+ was mainly present in the cytoplasm and at the cell membrane of tumor cells. The percentage of CD49f+ cells throughout the tumor was quite different for inter- and intratumors (Figure 2). As expected, the majority of tumors (197/312, 62.8%) contained no CD49f+ cells, whereas a minority of tumors contained CD49f+ cells at various frequencies. To better illustrate the distribution of CD49f+ cells in the tumors, we scored the tumors based on the percentage of CD49f+ cells relative to total cells, as previously reported [2].
To better understand the clinical significance of CD49f+ cells in tumors, we analyzed the association between CD49 positive tumors and clinical outcomes stratified by tumor histopathological characteristics, including age, WHO grade, nodal status, tumor size, HER2 status, ER status, distant metastasis, local recurrence, and recurrence (distant metastasis or/and local recurrence) after initial surgery, using t-tests. In this study, although no significant associations were identified between CD49f positivity in the tumors and most clinic characteristics (Figure 3A-H), we observe correlations between CD49f+ and disease recurrence (Figure 3I).
Since the presence of CD49f+ tumors was associated with disease recurrence, we analyzed the connection between this marker and patient survival using univariate survival analysis. First, we analyzed all of the patients (n=312) for DFS and OS. As shown in Figure 4A, patients bearing tumors with CD49f+ cells had shorter DFS after initial surgery (p=0.024). For example, 5 years after initial surgery, there was a nearly 20% DFS difference between the CD49 negative and positive groups (Figure 4A). However, no significant difference was observed in the OS analysis (p=0.235) (Figure 4B). To correct for treatment bias, we further excluded patients who did not receive chemotherapy (18 patients). For the rest of the 294 patients, we also found that tumors with CD49f+ cells were negatively associated with DFS (p=0.044) (Figure 4C). Again, we did not find any significant difference in the OS analysis (p=0.283) (Figure 4D).
To further confirm the previous results, we conducted a multivariate analysis using a Cox proportional hazard model. Prevalence of CD49f+ cells, age, WHO grade, tumor size, nodal status, ER, PR, and HER2 were included as covariates in the DFS and OS analysis. The analysis confirmed that CD49f+ cells in tumors are strong prognostic indicators for DFS (hazard ratio, 1.800; 95% confidence interval, 1.101–2.941; p=0.019). As expected, age and PR status were also found to be prognostic factors for DFS. WHO grade, tumor size, nodal status, PR, and HER2 were prognostic factors for OS (Table 2).
Our work suggests that mammary epithelial progenitors/stem cell-like cells can be used as biomarkers for prognostic purposes. By staining for CD49f+ cells in breast cancer tissues, we can differentiate patients who have a higher probability of distant metastasis and local recurrence that will lead to worse clinical outcomes. This simple strategy could potentially be used for prognosis and treatment decision purposes during conventional surgical pathology work.
In breast cancer, it is not initial tumor formation but rather metastasis and local recurrence that are the main causes of treatment failure and death [15]. Therefore, it is necessary to identify tumors with a high probability of distant metastasis and local recurrence to achieve personalized healthcare and develop more effective therapies. Numerous genes and steps have been identified as being involved in the process of distant metastasis. Briefly, in the initial step, signaling pathways that regulate cytoskeletal dynamics and the formation of cell-cell junctions are turned on in tumor cells, resulting in the migration of “activated tumor cells” into adjacent tissues [16]. After invading the adjacent tissues, the disseminated activated tumor cells may interact with blood or lymph vessels, penetrating the basement membrane and endothelial walls and entering the blood or lymph circulatory systems [17,18]. Under certain conditions, the circulating tumor cells attach to distant organs and proliferate to form another tumor at the distant site, leading to metastasis [19]. Then, the newly formed tumor on the distant organ also begins to proliferate, invading adjacent tissues, entering the blood or lymph, eventually leading to a second distant metastasis [1820].
Instead of screening for driver genes for tumor distant metastasis and local recurrence, we are more interested in searching for the cell sources that actively engage in dissemination, or in other words, we want to find the “activated tumor cells” that interact with blood vessels and lymph vessels. Since these “activated tumor cells” need to dedifferentiate and then differentiate into a mesenchymal state (epithelial–mesenchymal transition), we want to determine whether tumor cells with stem cell/progenitor-like properties are the cell source for distant metastasis and local recurrence.
It is well known that breast tumors are highly heterogeneous, with many different types of tumor cells [11]. However, some believe that rather than being simply a chaotic collection of cells, a specific cell subgroup has the ability to recapitulate the entire tumor, leading to tumor recurrence, contributing to tumor heterogeneity, resisting chemotherapy and radiation therapy, and excluding intracellular toxins, sharing many phenotypic similarities with adult tissue stem cells [21222324].
Because BCSCs can also promote the epithelial–mesenchymal transition [25], including downregulation of E-cadherin and cell-cell junctions [18], BCSCs are widely accepted as one of the cell types that contribute to distant metastasis. There are two models for BCSC action: (1) malignant cells accumulate mutations and acquire a BCSC phenotype through clonal evolution, or (2) cancer arises in normal MaSCs [26]. In the first model, cancer cells are believed to spread to other organs during the late stage of cancer. In contrast, in the latter model, metastasis can happen at very early stages, and it may be possible to predict the possibility of distant metastasis after initial mastectomy by identifying BCSCs in the tumor.
During regular surgical treatment, excision of the bulk tumor can remove or kill most of the tumor cells; however, BCSCs can remain after treatment, acting as a cell source for local recurrence and distant metastasis (Figure 5A). In contrast, targeting BCSCs during treatment in combination with regular treatment may lead to the complete eradication of tumor cells in the patient's body (Figure 5B).
CD49f (integrin α6) has been shown to play essential roles in development, cell differentiation, and cell fate determination, and it is involved in cell migration, adhesion, survival, polarity, growth, and death [2728]. In breast cancer, CD49f is reportedly involved in certain stem cell-like activities, such as CD29- and CD49f-mediated metastasis, and CD44 and CD49f positive cells mediate stem cell properties in basal-like breast cancer [29]. As the EpCAMlow/CD49f+ subpopulation is enriched for MaSC (bipotent) progenitors, whereas the EpCAM+/CD49f+ subpopulation is enriched for luminal progenitors[12], we proposed that CD49f+ cells in breast cancer tumors may possess certain stem cell/progenitor cell-like properties, making them useful as potential prognostic biomarkers. Our results indicated that by simply labeling CD49f positive cells in a tumor, we could easily identify patients at high risk of distant metastasis, local recurrence, and poor clinical outcome. When performed after the initial surgery, this strategy may help us to better predict the disease outcome, leading to more personalized treatment.
Because tumorigenesis involves many complex biological mechanisms, a single cell-based labeling tool may not be sufficient to identify all cells with tumorigenic potential. Therefore, further studies should focus on analyzing other BCSC markers for clinical significance and to determine their functional mechanisms.
In conclusion, breast cancers with high numbers of CD49f+ cancer cells were associated with an increased risk of disease recurrence after initial surgery and with a poor clinical outcome (decreased DFS). This detection strategy is a potentially useful tool for prognosis and treatment purposes during conventional surgical pathology work.
Figures and Tables
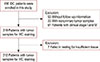 | Figure 1Flowchart showing the inclusion criteria used in this study.IDC=invasive ductal carcinoma; IHC=immunohistochemistry.
|
 | Figure 2Immunohistochemistry staining on tissue microarray (TMA) slides. Immunohistochemistry staining with primary antibodies against CD49f for all formalin-fixed paraffin-embedded TMA tissues was performed. The prevalence of CD49f positive cells was clearly different for inter- and intra-tumors. To quantify the prevalence of CD49f positive cells, we scored all tumors as either 0 (0% positive cells) (A), 1 (1%–10% positive cells) (B), 3 (51%–75% positive cells) (C), and 4 score (76%–100% positive cells) (D) (×200). |
 | Figure 3Prevalence of CD49f+ and histopathological characteristics. Analysis of 312 breast cancer patient tumor samples stained for CD49f and stratified for tumor age (A), WHO grade (low=grade 1 and 2, high=grade 3) (B), node status (C), tumor size (D), HER2 status (E), ER expression (F), distant metastasis (G), local recurrence (H), and disease recurrence (I). Tumors containing CD49f+ cells were associated with disease recurrence (distant metastasis or/and recurrence) (p=0.009).WHO=World Health Organization; HER2=human epidermal growth factor receptor 2; ER=estrogen receptor.
|
 | Figure 4Univariate survival analysis of CD49f+ with disease-free survival (DFS) and overall survival (OS) for total and chemotherapy-treated patients. Univariate survival analysis of CD49f+ cells with DFS and OS for total (A, B) and chemotherapy-treated patients (C, D). The presence of CD49f+ tumor cells had a significant negative association with DFS in both the total patient (A) and the chemotherapy-treated patient (C) groups. The present of CD49f+ tumor cells had no significant association with OS in the total patient (B) or the chemotherapy-treated patient (D) groups. |
 | Figure 5Identification of breast cancer stem cells (BCSCs) for treatment significance. (A) Although regular treatment involving bulk tumor excision can remove most of the tumor cells, BCSCs remain behind after treatment. These residual cells are the cellular source of recurrence and distant metastasis. (B) Targeting BCSCs along with conventional treatment of normal tumor cells may completely eradicate tumor cells in the patient. |
Table 1
Baseline clinical characteristics

Table 2
Multivariate Cox analysis of the histopathologic characteristics with CD49f+ cells

References
1. American Cancer Society. Cancer Treatment & Survivorship Fact & Figures 2012-2013. Atlanta: American Cancer Society;2012.
2. Ye F, Qiu Y, Li L, Yang L, Cheng F, Zhang H, et al. The presence of EpCAM(-)/CD49f(+) cells in breast cancer is associated with a poor clinical outcome. J Breast Cancer. 2015; 18:242–248.

3. Qiu Y, Pu T, Li L, Cheng F, Lu C, Sun L, et al. The expression of aldehyde dehydrogenase family in breast cancer. J Breast Cancer. 2014; 17:54–60.

4. Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004; 351:2817–2826.

5. Saghatchian M, Mook S, Pruneri G, Viale G, Glas AM, Guerin S, et al. Additional prognostic value of the 70-gene signature (MammaPrint(®)) among breast cancer patients with 4-9 positive lymph nodes. Breast. 2013; 22:682–690.

6. Sapino A, Roepman P, Linn SC, Snel MH, Delahaye LJ, van den Akker J, et al. MammaPrint molecular diagnostics on formalin-fixed, paraffin-embedded tissue. J Mol Diagn. 2014; 16:190–197.

7. Slodkowska EA, Ross JS. MammaPrint 70-gene signature: another milestone in personalized medical care for breast cancer patients. Expert Rev Mol Diagn. 2009; 9:417–422.

8. Bellocq JP, Luporsi E, Barrière J, Bonastre J, Chetritt J, Le Corroller AG, et al. uPA/PAI-1, Oncotype DX™, MammaPrint(®): prognosis and predictive values for clinical utility in breast cancer management. Ann Pathol. 2014; 34:349–351.

9. Ahmad S, Gupta S, Kumar R, Varshney GC, Raghava GP. Herceptin resistance database for understanding mechanism of resistance in breast cancer patients. Sci Rep. 2014; 4:4483.

10. Martin-Castillo B, Oliveras-Ferraros C, Vazquez-Martin A, Cufí S, Moreno JM, Corominas-Faja B, et al. Basal/HER2 breast carcinomas: integrating molecular taxonomy with cancer stem cell dynamics to predict primary resistance to trastuzumab (Herceptin). Cell Cycle. 2013; 12:225–245.
11. Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24-/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005; 11:1154–1159.
12. Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009; 15:907–913.

13. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010; 28:2784–2795.

14. Honeth G, Bendahl PO, Ringnér M, Saal LH, Gruvberger-Saal SK, Lövgren K, et al. The CD44+/CD24- phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008; 10:R53.
15. Sun B, Zhang S, Zhang D, Li Y, Zhao X, Luo Y, et al. Identification of metastasis-related proteins and their clinical relevance to triple-negative human breast cancer. Clin Cancer Res. 2008; 14:7050–7059.

17. Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003; 3:453–458.

18. Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011; 147:992–1009.

19. Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell. 2014; 158:1110–1122.

20. Hou JM, Krebs M, Ward T, Sloane R, Priest L, Hughes A, et al. Circulating tumor cells as a window on metastasis biology in lung cancer. Am J Pathol. 2011; 178:989–996.

21. Pfefferle AD, Spike BT, Wahl GM, Perou CM. Luminal progenitor and fetal mammary stem cell expression features predict breast tumor response to neoadjuvant chemotherapy. Breast Cancer Res Treat. 2015; 149:425–437.

22. Yang Y, Otte A, Hass R. Human mesenchymal stroma/stem cells exchange membrane proteins and alter functionality during interaction with different tumor cell lines. Stem Cells Dev. 2015; 24:1205–1222.

23. Zheng Z, Shao N, Weng H, Li W, Zhang J, Zhang L, et al. Correlation between epidermal growth factor receptor and tumor stem cell markers CD44/CD24 and their relationship with prognosis in breast invasive ductal carcinoma. Med Oncol. 2015; 32:275.

24. Pietras A, Katz AM, Ekström EJ, Wee B, Halliday JJ, Pitter KL, et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014; 14:357–369.

25. Huang Y, Yu P, Li W, Ren G, Roberts AI, Cao W, et al. p53 regulates mesenchymal stem cell-mediated tumor suppression in a tumor microenvironment through immune modulation. Oncogene. 2014; 33:3830–3838.

26. Badve S, Nakshatri H. Breast-cancer stem cells-beyond semantics. Lancet Oncol. 2012; 13:e43–e48.

27. Hwang R, Varner J. The role of integrins in tumor angiogenesis. Hematol Oncol Clin North Am. 2004; 18:991–1006.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download