INTRODUCTION
METHODS
Ethics statement
Mice and cell lines
Plasmid DNA construction
Fluorescence microscopy and flow cytometry analysis
Western blot analysis
Quantitative real-time PCR analysis
NO assay
Parasite growth and transfections
Extraction of genomic DNA and PCR confirmation of IP-10-egfp gene integration
RNA extraction and reverse transcription PCR
Immunization schedules
Histopathological studies
Sample preparation for arginase assay
Determination of arginase activity
Statistical analysis
RESULTS
Construction of recombinant L. tarentolae expressing IP-10-EGFP
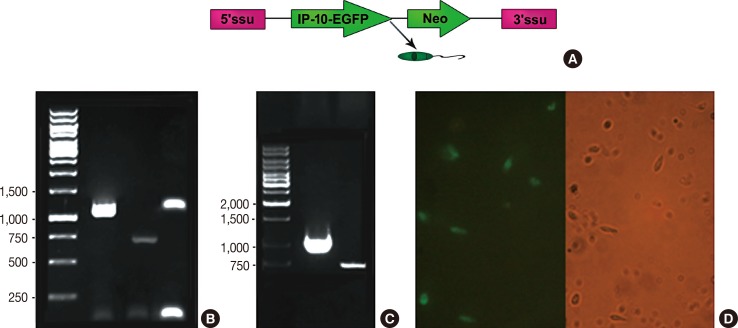 | Figure 1Interferon γ-induced protein 10-enhanced green fluorescent protein (IP-10-EGFP) integration and expression in Leishmania tarentolae strain. (A) Schematic presentation of egfp gene into ssu ribosomal DNA lucus. (B) Polymerase chain reaction (PCR) results using L. tarentolae genomic DNA as template. Columns 1 and 2 demonstrate the IP-10-EGFP and EGFP amplification, respectively. Column 3 is the PCR amplification of the integrated fragment using integration primers (F3001 and A1715) primers. (C) Confirmation of IP-10-EGFP expression in mRNA level. Amplification of IP-10-EGFP and EGFP fragments were demonstrated in column 1 and 2 using recombinant L. tarentolae cDNA as template, respectively. (D) Fluorescence microscopic images of recombinant L. tarentolae stably expressing (IP-10-EGFP) 48 hours postinfection; both bright field and fluorescence (left) are shown. |
In vitro expression of IP-10 EGFP in COS-7 transfected cells
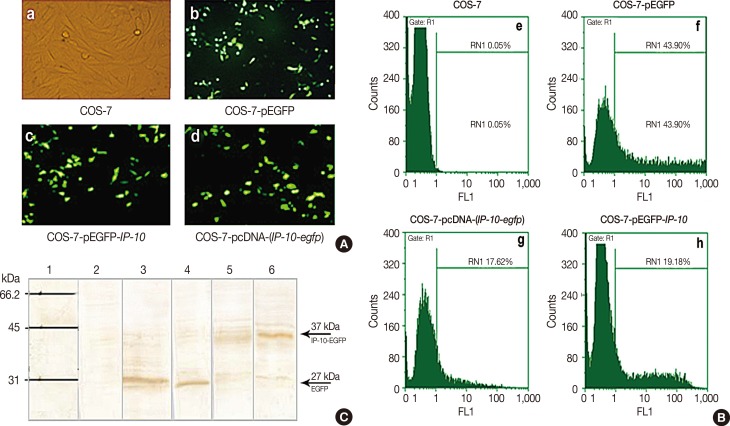 | Figure 2Confirmation of interferon γ-induced protein 10 (IP-10) expression in COS-7 transfected cells. (A) GFP expression was assessed by fluorescence microscopy after 48 hours. (a) Untreated COS-7, (b) COS-7 treated with pEGFP, (c) COS-7 treated with pEGFP-(IP-10), and (d) COS-7 treated with pcDNA-(IP-10-egfp). (B) GFP positive percentage of transfected parasite by flowcytometer after 48 hours. (e) Untreated COS-7, (f) COS-7 treated with pEGFP, (g) COS-7 treated with pcDNA-(IP-10-egfp), and (h) COS-7 treated with pEGP-IP-10. (C) Western blot analysis using anti-GFP antibody. Column 1 and 2 show marker and untreated COS-7 as negative control, respectively. The ~27 kDa band determines the GFP protein expression in Leishmania tarentolae-EGFP as positive control (column 3) and pEGFP-transfected COS-7 cells (column 4). IP-10-EGFP expression was confirmed by detecting ~37 kDa in pcDNA-(IP-10-egfp) (column 5) and pEGFP-(IP-10-EGFP) treated COS-7 cells (column 6).GFP=green fluorescent protein; EGFP=enhanced green fluorescent protein; R1=COS-7 cells; RN1=GFP positive COS-7; FL1=fluorescein isothiocyanate (FITC) detector.
|
Identification and verification of IP-10 protein
Quantification of IP-10 expression in pcDNA-(IP-10-egfp)-transfected COS-7 cells
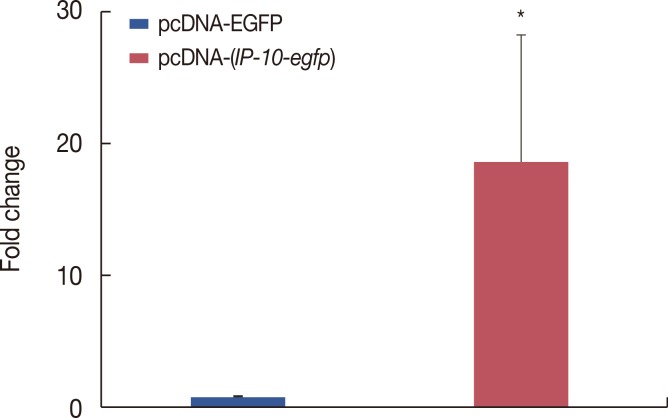 | Figure 3Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of interferon γ-induced protein 10 (IP-10) expression in pcDNA-(IP-10-egfp)-transfected COS-7 cells. qRT-PCR were performed 72 hours after transfection of pcDNA-egfp and pcDNA-(IP-10-egfp) into COS-7 cells. Data are presented as mean±SD (n=4).EGFP=enhanced green fluorescent protein.
*p<0.05.
|
NO induction in pcDNA-(IP-10-egfp)-transfected COS-7 cells
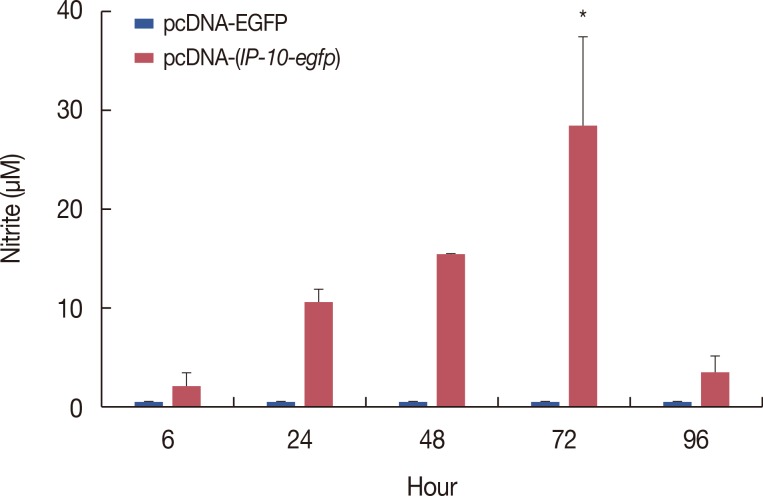 | Figure 4Effect of interferon γ-induced protein 10 (IP-10) on nitric oxide production. The supernatant of pcDNA-egfp and pcDNA-(IP-10-egfp)-transfected COS-7 cells were collected 6, 24, 48, 72, and 96 hours after transfection. Nitrate in the supernatant were measured with Griess reaction. Data are presented as mean±SD (n=4).EGFP=enhanced green fluorescent protein.
*p<0.05.
|
Effect of pcDNA-(IP-10-egfp) on tumor growth, tumor weight, and spleen weight
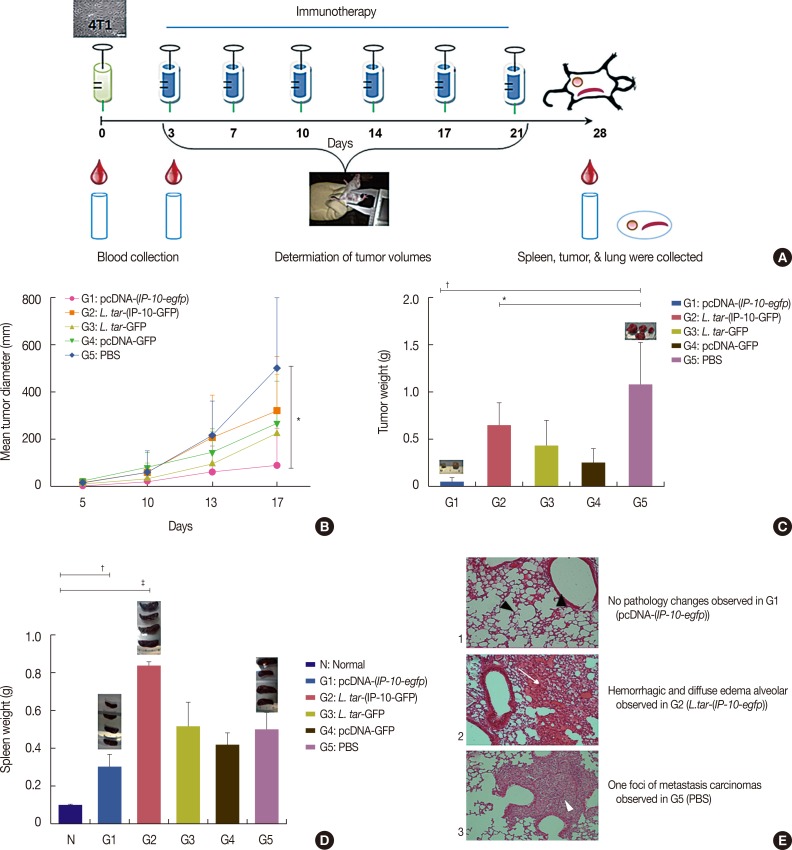 | Figure 5The effect of different therapeutic strategy on 4T1 tumor growth, spleen growth and lung histological sections. (A) Schematic representation of the immunotherapy schedule. BALB/c mice (n=10) were injected subcutaneously with 1×105 4T1 cells. Different immunization regimes were given intratumorally twice a week for 3 weeks. (B) Tumor volumes were determined two times per week. (C) Tumors were sectioned at day 28 and weight was measured among the five groups. (D) Spleens also were sectioned and weighted. Data are presented as mean±SD (n=4). (E) At day 28 lungs were removed, sectioned and stained with H&E. (1) Normal alveolar cavity with normal respiratory bronchioles (black arrowhead) and no significant pathologic changes in pcDNA-(IP-10-egfp) (G1). Alveolar septa have normal thickness with no congestion, edema or inflammation. (2) Hemorrhagic lung tissue in L. tarentolae-(IP-10-GFP) (G3) with diffused alveolar edema fluid in alveolar spaces (arrow). Alveolar septa show congestion. (3) One foci of metastatic carcinoma cells in field (white arrowhead) in G5. Arrow head shows sheet of metastatic carcinoma which encircle a respiratory bronchiole. The tumoral cells have high nuclear/cytoplasmic ratio, vascular pleomorphic nuclei and prominent nucleoli (H&E stain, ×100).IP-10-GFP=interferon γ-induced protein 10-green fluorescent protein; L. tar=Leishmania tarentolae; PBS=phosphate buffered saline.
*p<0.05; †p<0.001; ‡p<0.0001.
|
Histopathological studies in mice
Levels of arginase activity in isolated tumors and sera
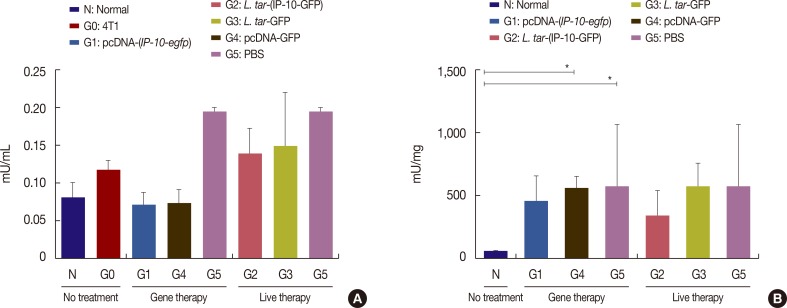 | Figure 6Arginase activity in tumor tissues and sera. Different immunization regimens were given to BALB/c mice (n=10) twice a week for 3 weeks, 3 days after 4T1 injection. (A) Arginase activity of different groups was measured in sera samples before 4T1 injection (N, normal), 3 days after injection 4T1 (G0, 4T1) and 23 days after injection of 4T1 (G1–G5). (B) Arginase activity in different groups was determined in tumor tissues 28 days after 4T1 injection. Data are representative of mean±SD (n=4).IP-10-GFP=interferon γ-induced protein 10-green fluorescent protein; L. tar=Leishmania tarentolae; PBS=phosphate buffered saline.
*p<0.05.
|




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download