INTRODUCTION
Breast cancer is composed of a heterogeneous group of cells that differ in morphology, marker expression, proliferative capacity, and tumorigenicity [
1]. Although many therapeutic treatments and prognostic factors have been identified for breast cancer, more accurate prognostic factors and ideal treatment modalities continue to be sought. Cancer stem cells (CSCs) are an emerging topic for breast cancer research. CSCs have the capacity for self-renewal, driving tumorigenicity, recurrence, metastasis, and the capacity to differentiate, albeit aberrantly, giving rise to a heterogeneous population of cancer cells [
23]. They are also believed to play a key role in resistance to chemotherapy and radiotherapy [
45].
The CSC theory was originally established for hematopoietic neoplasms and has greatly changed the concept of cancer therapy [
6]. Due to rapid development in biomolecular techniques, various new cell surface breast cancer markers have been discovered, such as aldehyde dehydrogenase 1 (ALDH1), CD133, and the presence of CD44 combined with the absence of CD24. However, expression of these CSC markers varied among different molecular subtypes of breast cancer and with different clinicopathologic features, such that each CSC population could have distinct clinical importance in different subgroups of breast cancers [
2]. ALDH1, which is a detoxification enzyme involved in catalyzing the oxidation of acetaldehydes produced from ethanol, has been suggested as another putative marker. High ALDH1 activity is characteristic of liver cancer and CSCs of breast and colon cancer [
57]. ALDH1 is associated with biologically aggressive phenotypes and worse clinical outcomes for breast cancer [
389]. CD133 also is considered a CSC marker in breast cancer, as well as in colorectal, brain, prostate, pancreatic, and gastric cancers [
10]. CD133, also known as Prominin 1, is a pentaspan transmembrane glycoprotein and was first described in hematopoietic stem cells [
11]. The biologic functions of CD133 in breast cancer are not completely established; however, CD133 may participate in tumor initiation, cellular migration, and vasculogenic mimicry [
12]. Overexpression of CD133 in triple-negative or node negative disease has been reported, and CD133 expression may help predict aggressive properties of breast cancer and determine optimal treatment more accurately [
1013]. Because of insufficient understanding of breast cancer CSCs, however, there is still debate regarding their exact biological functions, effects, and associations with clinicopathologic characteristics and prognosis in breast cancer. To more accurately estimate prognosis using CSCs, further studies of CSC markers are needed.
In this study, we evaluated the expression of two CSC markers, CD133 and ALDH1, and correlated their expression with clinicopathologic characteristics and prognosis in 291 patients with primary invasive breast cancer.
METHODS
Patients and samples
A total of 291 consecutive patients with invasive breast cancer (stage I, II, III) who underwent breast cancer surgery at Uijeongbu St. Mary's Hospital from 2005 to 2010 were included in this study. Clinicopathologic parameters were collected retrospectively from the Breast Cancer Patients Registry at Uijeongbu St. Mary's Hospital. Patients were staged according to the seventh edition of the breast cancer staging guidelines from the American Joint Committee on Cancer. All patients underwent resection: either lumpectomy or modified radical mastectomy. No patients received neoadjuvant chemotherapy. Ductal carcinoma
in situ and stage IV patients were excluded. Tissue samples were obtained from paraffin-embedded resected specimens used for histopathologic diagnosis. Clinicopathologic characteristics included age (over 35 years old or not), histological grade (the Scarff-Bloom-Richardson grading system [
14], 1, 2 vs. 3), TNM stage, and expression of estrogen receptor (ER; positivity ≥1%), progesterone receptor (PR; positivity ≥1%), and human epidermal growth factor receptor 2 (HER2). A recent study using the large nationwide Korean Breast Cancer Society registry suggests that an age of 35 years at the time of diagnosis is a reasonable cutoff point for defining a young age for the diagnosis of breast cancer [
1516]. In this study, patient age was grouped as <35 and ≥35 years old. A patient was considered to have HER2 negativity with a HercepTest (DAKO, Glostrup, Denmark) score less than 3+, and if the score were 2+, HER2 negativity was confirmed by fluorescence
in situ hybridization analysis with the PathVysion kit (Abbott-Vysis, Downers Grove, USA). Triple-negative (ER-, PR-, and HER2-) breast cancer (TNBC) and luminal (luminal A, B) versus nonluminal (basal-like, HER2-enriched) molecular subtypes were also included. Patients who underwent breast surgery were administered postoperative chemotherapy and radiotherapy.
Patient deaths were identified using both the death certificate data from the Korea National Statistical Office and hospital medical records. This study was approved by the Institutional Review Board at Catholic University College of Medicine of Korea (UC13SASI0121).
Immunohistochemistry
All patients had received breast cancer operations. Tissues samples from each patient were fixed in buffered formalin and embedded in paraffin. CD133 and ALDH1 immunohistochemical staining were performed on serial 4 µm tissue sections from formalin-fixed and paraffin-embedded human breast cancer tissues. Paraffin slides were deparaffinized in xylene three times for 10 minutes each and rehydrated through a graded ethanol series to distilled water before incubation for 10 minutes with 3% hydrogen peroxide in methanol to inhibit endogenous peroxidase activity. For antigen retrieval, slides were treated with 10 mM citrate buffer (pH 6.0) at 98℃ for 15 minutes in a microwave oven and allowed to cool for 1 hour at room temperature. After incubation for 10 minutes in a blocking solution (Histo-Plus kit; Zymed, San Francisco, USA) containing 10% normal serum in phosphate-buffered saline (PBS), sections were incubated at 4℃ overnight in a humidified chamber with rabbit polyclonal anti-CD133 antibody (diluted 1:200; Abnova, Taipei, Taiwan) and mouse monoclonal anti-ALDH1 antibody (diluted 1:100; Abcam, Cambridge, UK) as primary antibodies. A biotinylated secondary antibody (Histo-Plus kit) was used to detect primary antibodies, and slides were incubated for 10 minutes at room temperature. The sections were rinsed three times in PBS and incubated with streptavidin-horseradish peroxidase complex (Histo-Plus kit) for 10 minutes. Localized antigen was revealed using 3,3'-diaminobenzidine tetrahydrochloride as a chromongen, and the slide was counterstained with hematoxylin.
Immunohistochemical assessment
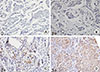
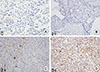
Microscopic analyses of CD133 and ALDH1 were assessed independently by two observers in a blinded manner. As in previous reports [
361113], scores were applied as follows: score 0, negative staining in all cells; score 1+, weakly positive or focally positive staining in <10% of the cells; score 2+, intermediate positive staining covering 10% to 50% of the cells; and score 3+, strongly positive staining, including >50% of the cells. For statistical analysis, CD133 and ALDH1 protein expression were considered positive when scores were ≥2 (
Figures 1,
2) [
613]. When discordance scores were obtained, two pathologists using a double-headed microscope reassessed the immunostained sections on a consensus basis, blinded to the clinicopathologic data.
Statistical analysis
Statistical analysis was performed with SAS version 9.3 (Statistical Analysis System, Cary, USA). The association between the expression of ALDH1 and CD133 and clinicopathological parameters was analyzed with the chi-square test. The Kaplan-Meier method was used to estimate disease-free survival (DFS) and overall survival (OS). DFS and OS were compared using a log-rank test. Events used to calculate DFS included all local, regional, or distant recurrences. The Cox regression model was used for multivariate analysis of prognostic factors. Test of independence (chi-square test) also was used to evaluate the association between CD133 and ALDH1. In all tests except the test of independence, a p-value <0.05 was considered to be statistically significant and, in the test of independence, a p-value >0.05 was considered to show independence between CD133 and ALDH1.
DISCUSSION
Over the past decade, CSCs have been studied and described in multiple types of cancers [
17]. The concept of CSCs was first identified in hematologic malignancies, and breast CSCs were first reported in 2003 by scientists from the University of Michigan Comprehensive Cancer Center, who concluded that only "a handful of CSCs" are required for growth, maintenance, and invasiveness of breast cancer [
18]. The CSC hypothesis has fundamental implications for cancer biology, in addition to clinical implications for cancer risk assessment, early detection, prognostication, and prevention. Development of more effective cancer therapies may require targeting the CSC populations [
3]. In other words, identification and characterization of CSCs could lead to development of directed and more effective treatments for cancer [
2].
CD133 and ALDH1 are emerging as important among the various newly-identified cell surface markers for breast cancer. Ginestier et al. [
3] and other investigators [
1618] have reported that ALDH1 is an adverse biomarker, associated with aggressive phenotypes and poor prognosis. CD133 expression also may increase the accuracy of predicting aggressive properties of breast cancer and determining optimal treatment [
10121920].
In the present study, we found that CD133 expression was significantly correlated with tumor size, LN metastasis, cancer stage, ER negativity, PR negativity, HER2 positivity, nonluminal subtype, TNBC, chemotherapy, and recurrence. Additionally, CD133 was associated with significant differences in DFS and OS. This finding suggests that CD133 expression is correlated with a number of adverse parameters that are traditionally associated with poor prognosis, and is an independent poor prognostic factor for invasive breast cancer. These results are very similar to Zhao et al.'s report [
13] that the expression of CD133 in 67 TNBC patients correlated with tumor size, LN status, and clinical stage, and was greatly associated with OS and DFS. In contrast, Ieni et al. [
19] reported that CD133 emerged as an independent prognostic variable for node negative breast cancer patients, but without a significant relationship with clinicopathologic parameters.
ALDH1 expression was also significantly correlated with tumor size, ER negativity, nonluminal subtype, TNBC, and recurrence. It showed a trend toward poor outcomes for both DFS and OS, but these were not statistically significant. However, the association with biologically aggressive phenotypes is similar to the study by Ginestier et al. [
3]. Morimoto et al. [
8] reported that ALDH1 expression was not significantly associated with prognosis, age, tumor size, LN status, or histological grade, but was significantly correlated with high ER negativity, PR negativity, HER2 positivity, high Ki-67, and high TOP2A expression. Resetkova et al. [
1] also reported that ALDH1 expression in breast cancer was associated with HER2 and triple-negative phenotypes but was not associated with DFS or OS in the cohorts treated with adjuvant or neoadjuvant chemotherapy. By contrast, other investigators reported that ALDH1 expression was significantly associated with DFS and OS [
136].
Although there was no correlation between the two CSC markers, our results also showed that CD133 expression is more widely associated than ALDH1 with adverse biomarkers and subtypes in invasive breast cancer. This finding was similar to the study by Aomatsu et al. [
10] evaluating CD133 and ALDH1 as potential surrogate markers for predicting sensitivity to neoadjuvant chemotherapy in breast cancer, in which only CD133 correlated with poor prognosis.
Combined expression of both CD133 and ALDH1 had statistical association with tumor size, cancer stage, histologic grade, ER negativity, HER2 positivity, nonluminal type, and TNBC. Furthermore, it also was significantly associated with poor DFS and OS.
Our present study showed that invasive breast cancers expressing CD133, ALDH1, or both antigens have biologically aggressive phenotypes, and expression of CD133 and the two together were more strongly associated with aggressive phenotypes than was ALDH1 expression. Expression of CD133 and both markers were also significantly associated with poor survival outcomes. These markers could potentially be used as independent prognostic factors for patients with invasive breast cancer. This study showed that CD133 or the combination of CD133 and ALDH1 may be clinically useful markers for identifying biologically aggressive breast cancer and predicting survival outcomes.
This study has several limitations. The first is an inadequate sample size. ALDH1 expression may actually portend worse clinical outcomes, but a larger sample size may be necessary to demonstrate such a relationship. The retrospective study design is a second limitation. Our study was conducted retrospectively, using patients who were treated with various adjuvant therapies that might affect prognosis and, as such, may have been subject to biases inherent to such study designs. Third, the short follow-up duration is another limitation, and a longer observation period is needed to evaluate prognosis. As such, further evaluations and studies are needed to overcome the limitations of our present study and to determine the exact biological functions of CSC markers in breast cancer. The final limitation is that there is no objective standard scoring system for reading the immunohistochemical staining of the CD133 and ALDH1 proteins.
In conclusion, our study showed that CD133 or the combination of CD133 and ALDH1 expression was more widely associated with adverse biomarkers and subtypes, compared with ALDH1 expression alone, of invasive breast cancer. These CSC markers were strongly associated with adverse biomarkers and subtypes of breast cancer, and these markers may have a predictive role and be helpful tools in the management of patients with invasive breast cancer.











 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download