Abstract
Since the introduction of the "cancer stem cell" theory, significant developments have been made in the understanding of cancer and the heterogenic structure of tumors. In 2003, with the isolation of cancer stem cells from the first solid tumor, breast cancer, and recognition of the tumorigenicity of these cells, this theory suggested that the main reason for therapy failure might be the presence of cancer stem cells. This review article describes breast cancer stem cell origin, the related cellular and molecular characteristics, signaling pathways, and therapy resistance mechanisms. The databases PubMed, SCOPUS, and Embase were explored, and articles published on these topics between 1992 and 2015 were investigated. It appears that this small subpopulation of cells, with the capacity for self-renewal and a high proliferation rate, originate from normal stem cells, are identified by specific markers such as CD44+/CD24-/low, and enhance a tumor's capacity for metastasis, invasion, and therapy resistance. Cancer stem cell characteristics depend on their interactions with their microenvironment as well as on the inducing factors and elements. Although uncertainties about breast cancer stem cells exist, many of researchers believe that cancer stem cells should be considered as possible therapeutic targets.
Breast cancer is the most prevalent type of cancer among women worldwide, representing 25% (1.7 million) of new cases and 15% (more than 0.5 million) of deaths from all cancers [1]. Different therapeutic methods/strategies are commonly employed, including local interventions (surgery/radiotherapy), and systemic treatments (chemo/hormonal therapy or targeted treatments) [2]. However, treated patients can suffer from disease relapse and metastasis [3] due to the presence of a subset of tumor cells known as breast cancer stem cells (BCSCs). BCSCs are a small cell population with unique characteristics such as self-renewal, high proliferation rate, and the ability to generate heterogenic lineages of cancer cells [4], attracting the attention of many researchers for their potential for use as a target for cancer therapy. In this article, the most important characteristics of BCSCs are reviewed.
Different theories have been proposed about the origin of BCSCs. One of them states that unsuitable regulation or mutations may lead to transformation of dormant normal stem cells to cancer stem cells (CSCs) [5]. According to another, the "misplacement somatic stem cell" theory, CSCs may originate from misplacement of somatic stem cells de novo [6]. Evidence shows that somatic cells can be considered the CSC origin. For example, Mintz et al. [7] indicated the teratogenic effect of somatic cells thorough injection of embryonic somatic cells into a mouse embryo aged 6 days. Several studies suggest that there are intratumoral lineages that have differentiated from common progenitor cells [8].
Surface markers, used for the isolation and identification of BCSCs, not only contribute to cell interactions, but also endow them with unique properties. For the first time in 2003, BCSCs were identified and isolated with the CD44+/CD24-/low Lin- phenotype [9]. Since then, the CD44+/CD24- phenotype has been used as a reliable phenotype for the isolation of BCSCs [10111213]. CD44 is a cell surface glycoprotein and specific receptor to hyaluronan. It is a key element for breast cancer adhesion, motion, migration, and invasion [14], and its interaction with osteopontin leads to tumor progression [15]. CD44 has an important role in cell proliferation and tumor angiogenesis [16]. CD24, another surface glycoprotein expressed at low levels, increases a tumor's ability to grow and metastasize [17]. Despite the growing list of CSC markers, some researchers do not consider these markers suitable for identifying CSCs. For example, one report shows that CD44+CD24- is not expressed in all breast cancer cell populations [18].
The other recently recognized marker, aldehyde dehydrogenase (ALDH) [19], consists of a family of cytosolic enzymes involved in the oxidation of intracellular aldehydes and oxidizes retinol to retinoic acid during the differentiation of rudimentary stem cells [20]. ALDH1, the dominant form of the enzyme in mammals, mediates the conversion of retinaldehydes to retinoic acid [21]. The other markers that have been used to identify BCSCs are CD133 [11] and a CD44+ CD49fhi CD133/2hi phenotype found in tumorigenic cells [22]. In vivo and in vitro studies have introduced CD49f [23] and CD61 [24] as BCSC markers as well (Table 1).
Notch, Hedgehog, and Wnt pathways have been implicated in resistance to therapy and an increased number of BCSCs during/after treatment. These pathways play key roles during embryonic development and adult tissue homeostasis [2526]. Dysregulation of the Notch and Hedgehog pathways, which are involved in normal stem cell self-renewal and differentiation, result in a BCSC phenotype in breast cancer cells [27]. The Wnt pathway plays a pivotal role in stem cell self-renewal and preservation of an undifferentiated state [28].
Hedgehog is an embryonic development organizer pathway that activates Gli1- and Ptch1-positive modulators of the hedgehog pathway, thereby leading to BCSC proliferation [29]. The Notch pathway is important to cell differentiation and connections during both embryogenesis and adulthood. It targets genes that lead to high proliferation and apoptosis inhibition in cancer cells [30]. Examples of transcription factors targeted include cyclinD1, c-myc, CDKN1A, and HES-related repressor protein. This pathway has been reported to act in BCSCs [31]. In addition to signaling pathways, transcriptional factors are significant, too. The main transcriptional factors Sox2, Oct4, and Nanog act as master regulators of pluripotency and maintain the undifferentiated state of cells [32]. Of basal-like breast carcinomas, 43% exhibit Sox2 expression, indicating a less differentiated phenotype [33]. In addition, there is evidence that Sox2 is expressed in derived spheres, those that have been generated from breast cancer tumors and cell lines [34].
Another member of the Sox family, Sox4, induces changes in the epithelial-mesenchymal transition (EMT) process, accompanied by an enhanced number of cells with a CD44+/CD24- phenotype and higher invasion and mobility of cancer cells in vivo and in vitro [35]. In addition, the role of protooncogenes and tumor suppressors is undeniable. They function to coordinately control stem cell self-renewal. For instance, excessive expression of c-myc, KLF4, Oct3/4, and Sox2 results in the differentiation of somatic cells into induced pluripotent stem cells [36]. Nevertheless, proteins encoded by these genes frequently act within less differentiated breast tumors and other tumors [3437]. Evidence suggests that the expression of Oct3/4, Nanog, and Sox2 are strongly associated with different CSCs, including BCSCs [383940].
Breast cancer therapy resistance is associated with the phenomenon of EMT and the reverse process, the mesenchymal-epithelial transition (MET) [41]. When BCSCs undergo EMT, the cells that survive chemo/hormonal therapy contain gene determinants similar to those of BCSCs and exhibit epithelial and mesenchymal markers (cytokeratin and vimentin, respectively) [4243]. Following chemo/hormonal therapy, EMT-like gene expression increases, indicating the role of epithelial-mesenchymal plasticity (EMP) in the development of resistance to cytotoxic drugs [424445]. EMP is defined as the ability of cells to undergo EMT and MET. During EMT, a series of changes take place, including the shutdown of transcription and down regulation of epithelial markers such as E-cadherin, and the appearance of mesenchymal markers such as vimentin, fibronectin, and N-cadherin. These changes lead to unstable structures and functions in these cells [46].
In addition, various growth factors, such as epidermal growth factor (EGF), tumor necrosis factor α (TNF-α), and transforming growth factor β (TGF-β), are expressed, which in turn activate mesenchymal transcriptional factors such as ZEB1, SNAIL1, SNAIL2, TWIST1, and TWIST 2. These factors inhibit ZO1, distinct claudins, and E-cadherin [47]. FOXC2, a transcriptional factor that is upregulated in tumors with high stem cell content, is the main marker of mesenchymal stem cells in breast cancer cell lines and of cells undergoing EMT [48]. High expression of SNAIL1, SNAIL2, and TWIST that are expressed during EMT leads to hyposensitivity to paclitaxel, adriamycin, and doxorubicin, respectively, indicating the role of EMP in BCSC therapy resistance [4449]. EMT also results in the resistance of the MCF7 cell line to tamoxifen [50].
The specific intratumoral condition location of cells is known as a microenvironment. The microenvironment of CSCs is referred to as a "niche," involving various factors that affect CSC properties. These factors include fibroblast stimuli, immune cells, autocrine signals, and extracellular matrix (ECM) components, as well as physical/chemical factors such as oxygen pressure, nutrients, and PH [51]. Tumor development also depends on cellular communication between different cell populations in a tumor niche [52]. The effects of the microenvironment on CSCs are discussed below.
A vast range of growth factors and cytokines released by tumor cells and cancer associated fibroblasts and immune cells ensure CSC survival and metastasis. Cytokines that are produced include interleukin (IL)-1, -6, and -8, CXCL12, CCL2, and growth factors such as platelet derived growth factor (PDGF), TGF-β, TNF-α, EGF, vascular endothelial growth factor, and FGF [53545556575859606162]. Stimuli that are applied in a microenvironment influence BCSC properties and therapy resistance. For example, IL-6 pathway activity and consequently STAT3/NF-Kβ expression result in trastuzumab resistance and an increased number of HER2 positive BCSCs. IL-6 receptor blockage leads to in vivo inhibition of metastasis and tumor growth [54].
Increased activity of the TGFβ pathway and expression of IL-8, induced by paclitaxel, enhance the cancer stem cell content of triple-negative breast tumors. Blockage of TGF-β type1 and 2 receptors thorough IL-8 inhibition prevents an increase in the number of BCSCs [55]. Moreover, IL-8 receptor (CXCR1) blocking agents, such as certain antibodies or repertaxin, target BCSCs selectively and prevent tumor formation in preclinical models [56].
CXCL12 activates the CXCR4 pathway that is necessary for stem cell survival; therefore, pathway disruption results in less drug resistance. In addition, CXCL12 promotes cancer cell growth through matrix metalloproteinase (MMP)-associated tissue remodeling [575859]. Under the influence of environmental factors (i.e., radiation), the microenvironment produces various types of growth factors, such as IL-1, CXCL12, TNF-α, TGF-β, PDGF, and MMPs. Growth factors facilitate tumor survival and regrowth and endothelial cell survival, and increase the generation rate of invasive CSCs [606162].
It is assumed that tissue oxygen pressure can influence CSCs. In oxygenated conditions, cells are active and have high migration and proliferation abilities. In low oxygen conditions (hypoxia) cells appear quiescent and radioresistant. The effect of hypoxia on the biology and physiology of tumors is paradoxical. Hypoxia causes CSC resistance in various ways. First, oxygen is known as a potential radiosensitizing factor. However, hypoxia induces greater radioresistance [63]. Hypoxia is associated with early relapse after radiotherapy, and higher oxygen levels improve patients' responses to treatment [64].
Second, the quiescent state and slower stem cell cycle due to hypoxia leads to greater chemo/radioresistance [65]. Hypoxia, in turn, can activate hypoxia inducible factor (HIF), serving as a CSC survival factor and EMT regulator [6667]. The most prominent HIFs, HIF1, and HIF2, are associated with the Wnt, Hedgehog, and Notch pathways [6869]. HIF1 serves as a key component for BCSC viability, as well as stem cell survival, metastasis, angiogenesis, EMT, and radio/chemo resistance [70]. Enhanced HIF-1α indicates a poor outcome, metastasis, and early relapse of breast cancer patients [71].
BCSC therapy resistance mechanisms are various, but the most significant of these are described below (Figure 1).
The ATP-binding cassette (ABC) family of membrane proteins is commonly involved in the transport of compounds and small molecules out of the cell. ABC transporters play an important role in the establishment of chemical homeostasis and cell survival in unfavorable conditions within a broad range of environments, including normal stem cells, the placenta, epithelial cells of the digestive system, and endothelial cells that form the blood-brain barrier [72]. In addition to physiological functions, ABC transporters have been reported to be important in multidrug resistance (MDR) in various cancers [73].
Among the 49 known ABC transporters, three of them have been shown to be MDR regulators, including glycoprotein P (P-gP), multidrug resistance protein 1 (MDR1), and breast cancer resistance protein [74]. These transporters are highly expressed in different cancers, including breast cancer, resulting in cytotoxic drugs being carried out of cells using ATP, and subtoxic drug doses maintained within a cell [75].
Also, overlap among transporters with a vast range of substrates provide tumor drug resistance to a large group of chemotherapy drugs, including antimetabolites, topoisomerase inhibitors, and taxanes, as well as tyrosine kinase inhibitor drugs, such as erlotinib, imatinib, gefitinib, nilotinib, and sorafenib, that serve as molecular targeting drugs [7476]. Studies have shown that preventing ATP-binding cassette G member 2 (ABCG2) transporter activity using Ko143 (fumitremorgin-type indolyl diketopiperazine) and phosphodiesterase-5 inhibitors can improve the performance of anticancer drugs [7778].
ALDH is an enzyme family involved in the oxidation of intracellular aldehydes to carboxylic acids, retinoic acid, and γ-amino butyric acid (GABA) biosynthesis, with a significant role in the survival and differentiation of normal and CSCs [7980]. ALDH induces CSC radioresistance both through direct removal of oxygen radicals that are produced and indirect production of an antioxidant compound, nicotinamide adenine dinucleotide (phosphate) [81].
In addition, ALDH1 seems to be associated with breast cancer malignancy, metastasis, and invasion [82]. High activity of ALDH1 and 3 isoforms enables the cells to metabolize cyclophosphamide and analogues such as ifosfamide, mafosfamide, and 4-hydroperoxy cyclophosphamide, and to detoxify aldophosphamide (an intermediated product) to carboxyphosphamide [8384]. ALDH+ cells not only exhibit paclitaxel and epirubicin resistance but also increase in number after chemotherapy [85].
Radiotherapy and various types of chemotherapy drugs damage DNA through various mechanisms, such as DNA synthesis inhibition (methotrexate), topoisomerase inhibitory action (daunorubicin, doxorubicin), and DNA cross-link formation (carboplatin, cisplatin, oxaliplatin), in which DNA repair failure results in cell death [86]. CSCs possess DNA repair and reactive oxygen species (ROS) scavenging mechanisms [25]. Nevertheless, DNA damage and repair mechanisms are generally different. In addition, DNA damage as an inducing factor may activate checkpoint mechanisms. DNA repair is proposed to be acting in various types of tumors, including human and mouse breast tumors [8788]. The most lethal type of DNA damage is a double-strand break, which is repaired in two ways [89].
Homology-directed recombination (HDR) is an error free repair mechanism. The recombination steps (catalyzed by distinct enzymes) include 3'-5' resegmentation of the two ends of DNA, formation of single-strand DNA at the 3' end, assembly of RAD51 filaments (a protein family contributing in the repair of DNA double-stand breaks), and finally repair synthesis by annealing at the end of the double-strand break [9091]. Since this method requires intact sister chromatids, HDR repair occurs only during the S and G2 phases of the cell cycle.
Unlike HDR repair, nonhomologous end joining (NHEJ) introduces error. In this process, the KU70/KU80 proteins join the ends of DNA strands. In addition, nucleases, polymerases, DNA-dependent protein kinases, and ligases participate in the NHEJ repair process [91].
Regarding DNA damage-activated checkpoint mechanisms, two pathways are noticeable. ATR-Chk1 and ATM-Chk2 are kinase-signaling pathways activated by double-strand and single-strand DNA breaks, respectively. DNA damage checkpoint signaling makes DNA repair possible thorough cell cycle inhibition [92]. The ATM/ChK2 activating pathway has been proposed to be associated with BCSCs radioresistance, and the existence of the ATM inhibitor and radiosensitivity enhancer, KU55933, suggests that the ATM pathway is an appropriate target for eliminating radioresistance in BCSCs [93].
Different levels of oxygen are essential to promoting intracellular reactions. ROS, active radicals that are produced during oxygen metabolism, participate in the regulation of physiologic events of the cell, such as proliferation, migration, wound healing, and angiogenesis [94]. Excess amounts of ROS produced in response to radiation, followed by interactions with cell components like DNA, proteins, and lipids, induce cell death [95].
Both normal and cancer cells establish an equilibrium between production and depletion of ROS using compounds such as catalase, glutathione peroxidase, superoxide dismutase, and thioredoxin [96]. BCSCs have specific mechanisms to guard against the genotoxic effects of ROS, including more effective ROS scavenging and lower levels of ROS production. In addition, genes encoding the proteins superoxide dismutase, catalase, and glutathione peroxidase enzymes, all of which are involved in ROS scavenging, are upregulated in BCSCs significantly. The importance of ROS scavenging appears when tumor treatment with buthionine sulfoximine (BSO) leads to lower radioresistance with decreased clonogenic potential of CSCs. BSO seems to promote this process by inhibiting glutamate cystein ligase [97].
Metastasis is a complicated process that begins with the migration of cancer cells from the primary tumor and is followed by regional invasion and entry to the circulatory system. Eventually, metastasis terminates with the arrest of cells at a secondary site, extravasion, and colonization. Colonization is not merely a single term; it involves a series of events, including survival of cancer cells until they enter the tissue, the formation of micrometastases, a latency phase, regrowth from latent micrometastases, progressive tumor growth surpassing host tissue growth, and recirculation and formation of tertiary lesions in the same or different organs [98].
The most common site for breast cancer metastasis is bone, but other organs such as the lung and liver are also involved [99]. Hyaluronan and osteopontin, the major components of breast cancer target tissues (bone, brain, liver, and lung), serve as specific ligands for CD44 [100]. Osteopontin, which is expressed in different tissues, contributes to cell adhesion and immune responses [101102]. Breast cancer metastatic cells attach to the bone marrow endothelium via osteopontin [103]. Osteopontin is also associated with a higher incidence of tumor metastasis and invasion [104]. Highly expressed osteopontin, IL-1, and CXCR4 enhance the metastatic potential of breast cancer cells [105].
In addition to osteopontin, one of the protein components of ECM, tenascin C, enhances the efficiency of signaling pathways Wnt and Notch, stabilizing breast cancer-initiating tumor cells in the lungs [106]. Breast cancer cells exhibit high Src tyrosine kinase activity, leading to P13K-Akt pathway sensitivity associated with cell survival. The sensitized P13K-Akt pathway, in turn, is activated by CXCL12 and IGF1, which are released from the bone marrow stroma [107].
Vascular cell adhesion molecule 1 (V-CAM1) is highly expressed in breast cancer cells, sensitizing the P13K-Akt pathway to external signals. Moreover, α4β1 integrins belonging to tumor-associated macrophages activate ezrin (adaptor protein for P13K and Akt) via V-CAM1 [108]. MMP1, which facilitates metastasis to the brain, has a high expression in cells that metastasize to brain. MMP1 breaks down occludin and claudin, the main components of the blood-brain barrier. Also, cyclooxygenase2 (COX2) upregulation induces prostaglandins that promote MMP1 expression directly. Astrocytes that have been activated by COX2 and prostaglandins produce chemokine ligand 7 (CCL7), which is involved in the progression of tumor-initiating cells in the brain [109].
CSCs may be subjected to unfavorable conditions, such as hypoxia, loss of nutrients, or toxic drugs, in their microenvironments. To resist these conditions, CSCs possess various catabolic processes to maintain their viability and metabolic homeostasis. The main mechanism for homeostasis maintenance is autophagy [110]. During autophagy, proteins or organelles form autophagosome-joining lysosomes that become autophagolysosomes. After this, lysosomal enzymes such as cathepsins break down their contents to return the energy or amino acids to the cells [111].
Autophagy failure is associated with muscle atrophy, degeneration of the nervous system, and a broad range of cancers [112]. In investigations of the role of autophagy in cancer, a dual function has been observed. On the one hand, it prevents the accumulation of damaged proteins and organelles, serving as a tumor suppressor, and on the other hand, it acts as a tumor enhancer with hypoxia or the loss of nutrients. Cell stress or increased metabolic requirements force tumor cells to activate autophagy [113].
Radiotherapy suppresses breast, prostate, glioblastoma, and colon cancers by enhancing autophagy hyperactivity of cancer cells and autophagy-induced cell death [114115]. In addition, D-cyclovirobuxine induces autophagy-dependent death in the MCF7 cell line [116]. Nevertheless, the main role of autophagy is maintaining of tumor cells in stressful conditions. Autophagy is a mechanism for preserving cell integrity under cytotoxicity, metabolic stress, or radiation-induced disruptions [114117118119].
Accordingly, loss of autophagy results in increased DNA damage and chromosomal disruption in cancer cells during stress [117118]. Autophagy inhibition causes higher radiosensitivity of breast, prostate, glioblastoma, and colon cancers [114119]. Studies have shown that autophagy markers LC3B, Atg5, and Atg12 are involved in autophagosome formation, and are expressed at high levels in breast cancer stem-like quiescent cells [120].
Autophagy inhibition prevents the invasion of breast cancer progenitor cells, as well as spheroid and xenograft tumor formation. Autophagy activity seems to be significantly higher in mammospheres than in a control group [121]. Moreover, autophagy has been reported to be higher in an ALDH+ population of no special type breast cancer [122]. Chloroquine can inhibit autophagy through the prevention of lysosomal mass breakdown. It also interferes with E-cadherin endocytosis, thereby inhibiting EMT [123].
DNA methylation is a process in which methyl groups are added to the 5' end of cytosine residues in the guanosine cap. In mammals, DNA methylation is accomplished by three types of DNA methyl transferase enzymes (DNMT1, 3a, and 3b). DNMT3a and DNMT3b serve to regulate the methylation patterns of genes [124]; therefore, they play a key role in the regulation of stem cell properties by de novo methylation [125]. DNMT3b seems to play an even more significant role in this respect, inducing aberrant DNA methylation patterns and maintaining the undifferentiated state of CSCs [126]. Exploring 68 methylated regions of BCSCs, hypomethylation pattern was reported to be more common in these regions than in nonstem cells, and to be associated with poor prognosis [127].
Micro RNAs (miRNAs) are noncoding regulatory RNAs contributing to posttranscriptional regulation of gene expression. miRNAs result in mRNA degradation through the inhibition of ribosome activity, mRNA translation, and deletion of the 3' end of the poly A tail and the 5' end of the guanosine cap [128]. miRNAs restore stem cell characteristics to those of normal and cancer stem cells; hence, their dysregulation is associated with tumorigenicity.
Several examples of the involvement of miRNA in tumorigenicity have been reported. miR21 is an oncogene miRNA that is involved in breast cancer EMT and maintains stem cell-like characteristics [129]. miR200c suppression appears to act in BCSC tumorigenicity [130]. miR9 is associated with a BCSC phenotype and EMT state [131]. miR203 loss is observed in the majority of BCSCs. Finally, miR203 re-expression induces differentiation and suppression of stem and mesenchymal cell properties [132].
In recent years, the cancer stem cell theory has led to new insights into cancer treatment. Among different types of cancers, breast cancer has received the most attention due to the number of people impacted. The isolation of BCSCs from a solid tumor, along with an extensive understanding of cellular, molecular, and signaling pathway mechanisms, enforce the belief that therapy failure and therapy resistance are due to the presence of cancer stem cells, a subpopulation that has high proliferative and metastatic potential and that can act as a reservoir for tumorigenicity.
BCSC function is remarkable in several aspects. First, this population is resistant to conventional therapies due to enhanced membrane transport by specific protein transporters, specific mechanisms of DNA repair, and ROS scavenging systems, and the ability to detoxify cytotoxic drugs. Second, transcriptional factors, signaling pathways, and tumor suppressor genes act to maintain and amplify a state of stemness. Third, extensive interactions among cancer stem cells, their microenvironments, and other cells present initiate a cascade of growth factors and inducing elements, which in turn influence cancer stem cell function. More studies are needed to investigate each of these aspects of BCSCs.
Figures and Tables
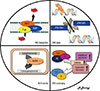 | Figure 1The most possible mechanisms of therapy resistance in breast cancer stem cells.MDR1=multidrug resistance protein 1; P-gp =glycoprotein P; BCRP=breast cancer resistance protein; ABC=ATP-binding cassette; ALDH1=aldehyde dehydrogenase 1; ROS=reactive oxygen species.
|
Table 1
Surface markers used to isolate the breast cancer stem cells

| Authors | Marker |
|---|---|
| Al-Hajj et al. (2003) [9] | CD44+/CD24-/low Lin- |
| Meyer et al. (2010) [22] | CD44+ CD49fhi CD133/2hi |
| Ginestier et al. (2007) [19] | ALDH1 |
| Wright et al. (2008) [11] | CD44+/CD24- CD133 |
| Cariati et al. (2008) [23] | CD49f |
| Vaillant et al. (2008) [24] | CD61 |
Notes
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65:87–108.

3. Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005; 365:1687–1717.
4. Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells: perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006; 66:9339–9344.

5. Hartwig FP, Nedel F, Collares T, Tarquinio SB, Nör JE, Demarco FF. Oncogenic somatic events in tissue-specific stem cells: a role in cancer recurrence? Ageing Res Rev. 2014; 13:100–106.

6. Wang RA, Li ZS, Zhang HZ, Zheng PJ, Li QL, Shi JG, et al. Invasive cancers are not necessarily from preformed in situ tumours: an alternative way of carcinogenesis from misplaced stem cells. J Cell Mol Med. 2013; 17:921–926.

7. Mintz B, Cronmiller C, Custer RP. Somatic cell origin of teratocarcinomas. Proc Natl Acad Sci U S A. 1978; 75:2834–2838.

8. Gorden BH, Kim JH, Sarver AL, Frantz AM, Breen M, Lindblad-Toh K, et al. Identification of three molecular and functional subtypes in canine hemangiosarcoma through gene expression profiling and progenitor cell characterization. Am J Pathol. 2014; 184:985–995.

9. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003; 100:3983–3988.

10. Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005; 65:5506–5511.

11. Wright MH, Calcagno AM, Salcido CD, Carlson MD, Ambudkar SV, Varticovski L. BRCA1 breast tumors contain distinct CD44+/CD24- and CD133+ cells with cancer stem cell characteristics. Breast Cancer Res. 2008; 10:R10.
12. Perrone G, Gaeta LM, Zagami M, Nasorri F, Coppola R, Borzomati D, et al. In situ identification of CD44+/CD24- cancer cells in primary human breast carcinomas. PLoS One. 2012; 7:e43110.

13. Wang LB, He YQ, Wu LG, Chen DM, Fan H, Jia W. Isolation and characterization of human breast tumor stem cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2012; 28:1261–1264.
14. Herrera-Gayol A, Jothy S. Adhesion proteins in the biology of breast cancer: contribution of CD44. Exp Mol Pathol. 1999; 66:149–156.

15. Rangaswami H, Bulbule A, Kundu GC. Osteopontin: role in cell signaling and cancer progression. Trends Cell Biol. 2006; 16:79–87.

16. Götte M, Yip GW. Heparanase, hyaluronan, and CD44 in cancers: a breast carcinoma perspective. Cancer Res. 2006; 66:10233–10237.

17. Schabath H, Runz S, Joumaa S, Altevogt P. CD24 affects CXCR4 function in pre-B lymphocytes and breast carcinoma cells. J Cell Sci. 2006; 119(Pt 2):314–325.

18. Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells: what challenges do they pose? Nat Rev Drug Discov. 2014; 13:497–512.

19. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007; 1:555–567.

20. Balicki D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell. 2007; 1:485–487.

21. Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, et al. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci U S A. 2006; 103:11707–11712.

22. Meyer MJ, Fleming JM, Lin AF, Hussnain SA, Ginsburg E, Vonderhaar BK. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010; 70:4624–4633.

23. Cariati M, Naderi A, Brown JP, Smalley MJ, Pinder SE, Caldas C, et al. Alpha-6 integrin is necessary for the tumourigenicity of a stem celllike subpopulation within the MCF7 breast cancer cell line. Int J Cancer. 2008; 122:298–304.

24. Vaillant F, Asselin-Labat ML, Shackleton M, Forrest NC, Lindeman GJ, Visvader JE. The mammary progenitor marker CD61/beta3 integrin identifies cancer stem cells in mouse models of mammary tumorigenesis. Cancer Res. 2008; 68:7711–7717.

25. Peitzsch C, Kurth I, Kunz-Schughart L, Baumann M, Dubrovska A. Discovery of the cancer stem cell related determinants of radioresistance. Radiother Oncol. 2013; 108:378–387.

26. Karamboulas C, Ailles L. Developmental signaling pathways in cancer stem cells of solid tumors. Biochim Biophys Acta. 2013; 1830:2481–2495.

27. Muñoz P, Iliou MS, Esteller M. Epigenetic alterations involved in cancer stem cell reprogramming. Mol Oncol. 2012; 6:620–636.

28. Ling L, Nurcombe V, Cool SM. Wnt signaling controls the fate of mesenchymal stem cells. Gene. 2009; 433:1–7.

29. Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006; 66:6063–6071.

30. Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008; 27:5124–5131.

31. Farnie G, Clarke RB. Mammary stem cells and breast cancer: role of Notch signalling. Stem Cell Rev. 2007; 3:169–175.

32. Yamanaka S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008; 41:Suppl 1. 51–56.

33. Rodriguez-Pinilla SM, Sarrio D, Moreno-Bueno G, Rodriguez-Gil Y, Martinez MA, Hernandez L, et al. Sox2: a possible driver of the basal-like phenotype in sporadic breast cancer. Mod Pathol. 2007; 20:474–481.

34. Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, et al. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012; 31:1354–1365.

35. Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012; 72:4597–4608.

36. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861–872.

37. Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008; 40:499–507.

38. Stolzenburg S, Rots MG, Beltran AS, Rivenbark AG, Yuan X, Qian H, et al. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012; 40:6725–6740.

39. Lu X, Mazur SJ, Lin T, Appella E, Xu Y. The pluripotency factor nanog promotes breast cancer tumorigenesis and metastasis. Oncogene. 2014; 33:2655–2664.

40. Luo W, Li S, Peng B, Ye Y, Deng X, Yao K. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS One. 2013; 8:e56324.

41. Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010; 29:4741–4751.

42. Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009; 106:13820–13825.

43. Creighton CJ, Chang JC, Rosen JM. Epithelial-mesenchymal transition (EMT) in tumor-initiating cells and its clinical implications in breast cancer. J Mammary Gland Biol Neoplasia. 2010; 15:253–260.

44. Li QQ, Xu JD, Wang WJ, Cao XX, Chen Q, Tang F, et al. Twist1-mediated adriamycin-induced epithelial-mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clin Cancer Res. 2009; 15:2657–2665.

45. Farmer P, Bonnefoi H, Anderle P, Cameron D, Wirapati P, Becette V, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009; 15:68–74.

46. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006; 7:131–142.

47. Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, et al. Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J Cell Physiol. 2007; 213:374–383.

48. Hollier BG, Tinnirello AA, Werden SJ, Evans KW, Taube JH, Sarkar TR, et al. FOXC2 expression links epithelial-mesenchymal transition and stem cell properties in breast cancer. Cancer Res. 2013; 73:1981–1992.

49. Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, Wang LH. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007; 67:1979–1987.

50. Hiscox S, Jiang WG, Obermeier K, Taylor K, Morgan L, Burmi R, et al. Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of beta-catenin phosphorylation. Int J Cancer. 2006; 118:290–301.

51. Borovski T, De Sousa E Melo F, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res. 2011; 71:634–639.
52. Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: attacking cancer's inflammatory roots. Clin Cancer Res. 2011; 17:6125–6129.

53. Brooks MD, Wicha MS. Tumor twitter: cellular communication in the breast cancer stem cell niche. Cancer Discov. 2015; 5:469–471.

54. Korkaya H, Kim GI, Davis A, Malik F, Henry NL, Ithimakin S, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012; 47:570–584.

55. Bhola NE, Balko JM, Dugger TC, Kuba MG, Sánchez V, Sanders M, et al. TGF-beta inhibition enhances chemotherapy action against triplenegative breast cancer. J Clin Invest. 2013; 123:1348–1358.

56. Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, et al. CXCR1 blockade selectively targets human breast cancer stem cells in vitro and in xenografts. J Clin Invest. 2010; 120:485–497.

57. Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009; 113:4341–4351.

58. Zeng Z, Samudio IJ, Munsell M, An J, Huang Z, Estey E, et al. Inhibition of CXCR4 with the novel RCP168 peptide overcomes stromamediated chemoresistance in chronic and acute leukemias. Mol Cancer Ther. 2006; 5:3113–3121.

59. Dubrovska A, Elliott J, Salamone RJ, Telegeev GD, Stakhovsky AE, Schepotin IB, et al. CXCR4 expression in prostate cancer progenitor cells. PLoS One. 2012; 7:e31226.

60. Kuonen F, Secondini C, Rüegg C. Molecular pathways: emerging pathways mediating growth, invasion, and metastasis of tumors progressing in an irradiated microenvironment. Clin Cancer Res. 2012; 18:5196–5202.

61. Chung YL, Jian JJ, Cheng SH, Tsai SY, Chuang VP, Soong T, et al. Sublethal irradiation induces vascular endothelial growth factor and promotes growth of hepatoma cells: implications for radiotherapy of hepatocellular carcinoma. Clin Cancer Res. 2006; 12:2706–2715.

62. Sofia Vala I, Martins LR, Imaizumi N, Nunes RJ, Rino J, Kuonen F, et al. Low doses of ionizing radiation promote tumor growth and metastasis by enhancing angiogenesis. PLoS One. 2010; 5:e11222.

63. Wardman P. Chemical radiosensitizers for use in radiotherapy. Clin Oncol (R Coll Radiol). 2007; 19:397–417.

64. Batchelor TT, Gerstner ER, Emblem KE, Duda DG, Kalpathy-Cramer J, Snuderl M, et al. Improved tumor oxygenation and survival in glioblastoma patients who show increased blood perfusion after cediranib and chemoradiation. Proc Natl Acad Sci U S A. 2013; 110:19059–19064.

65. Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010; 140:62–73.

66. Cho KH, Choi MJ, Jeong KJ, Kim JJ, Hwang MH, Shin SC, et al. A ROS/STAT3/HIF-1alpha signaling cascade mediates EGF-induced TWIST1 expression and prostate cancer cell invasion. Prostate. 2014; 74:528–536.

67. Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, Dubrovska A. Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther. 2013; 6:1347–1361.
68. Qiang L, Wu T, Zhang HW, Lu N, Hu R, Wang YJ, et al. HIF-1alpha is critical for hypoxia-mediated maintenance of glioblastoma stem cells by activating Notch signaling pathway. Cell Death Differ. 2012; 19:284–294.

69. Choi H, Chun YS, Kim TY, Park JW. HIF-2alpha enhances beta-catenin/TCF-driven transcription by interacting with beta-catenin. Cancer Res. 2010; 70:10101–10111.

70. Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012; 33:207–214.

71. Generali D, Berruti A, Brizzi MP, Campo L, Bonardi S, Wigfield S, et al. Hypoxia-inducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res. 2006; 12:4562–4568.

72. DeGorter MK, Xia CQ, Yang JJ, Kim RB. Drug transporters in drug efficacy and toxicity. Annu Rev Pharmacol Toxicol. 2012; 52:249–273.

73. Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002; 2:48–58.

74. Vasiliou V, Vasiliou K, Nebert DW. Human ATP-binding cassette (ABC) transporter family. Hum Genomics. 2009; 3:281–290.

75. Leonard GD, Fojo T, Bates SE. The role of ABC transporters in clinical practice. Oncologist. 2003; 8:411–424.

76. Huang WC, Hsieh YL, Hung CM, Chien PH, Chien YF, Chen LC, et al. BCRP/ABCG2 inhibition sensitizes hepatocellular carcinoma cells to sorafenib. PLoS One. 2013; 8:e83627.

77. Lecerf-Schmidt F, Peres B, Valdameri G, Gauthier C, Winter E, Payen L, et al. ABCG2: recent discovery of potent and highly selective inhibitors. Future Med Chem. 2013; 5:1037–1045.

78. Stacy AE, Jansson PJ, Richardson DR. Molecular pharmacology of ABCG2 and its role in chemoresistance. Mol Pharmacol. 2013; 84:655–669.

79. Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev. 2011; 7:292–306.

80. Young SZ, Bordey A. GABA's control of stem and cancer cell proliferation in adult neural and peripheral niches. Physiology (Bethesda). 2009; 24:171–185.

81. Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, et al. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013; 56:89–101.

82. Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, et al. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009; 13:2236–2252.

83. Magni M, Shammah S, Schiró R, Mellado W, Dalla-Favera R, Gianni AM. Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood. 1996; 87:1097–1103.

84. Parajuli B, Fishel ML, Hurley TD. Selective ALDH3A1 inhibition by benzimidazole analogues increase mafosfamide sensitivity in cancer cells. J Med Chem. 2014; 57:449–461.

85. Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, et al. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009; 15:4234–4241.

86. Cheung-Ong K, Giaever G, Nislow C. DNA-damaging agents in cancer chemotherapy: serendipity and chemical biology. Chem Biol. 2013; 20:648–659.

87. Al-Assar O, Mantoni T, Lunardi S, Kingham G, Helleday T, Brunner TB. Breast cancer stem-like cells show dominant homologous recombination due to a larger S-G2 fraction. Cancer Biol Ther. 2011; 11:1028–1035.

88. Kim SY, Rhee JG, Song X, Prochownik EV, Spitz DR, Lee YJ. Breast cancer stem cell-like cells are more sensitive to ionizing radiation than non-stem cells: role of ATM. PLoS One. 2012; 7:e50423.

89. Brandsma I, Gent DC. Pathway choice in DNA double strand break repair: observations of a balancing act. Genome Integr. 2012; 3:9.

90. Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb Perspect Biol. 2013; 5:a012740.

93. Yin H, Glass J. The phenotypic radiation resistance of CD44+/CD24(- or low) breast cancer cells is mediated through the enhanced activation of ATM signaling. PLoS One. 2011; 6:e24080.
94. Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012; 48:158–167.

95. Cook JA, Gius D, Wink DA, Krishna MC, Russo A, Mitchell JB. Oxidative stress, redox, and the tumor microenvironment. Semin Radiat Oncol. 2004; 14:259–266.

96. Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009; 8:579–591.

97. Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009; 458:780–783.

99. Del Mastro L, Clavarezza M, Venturini M. Reducing the risk of distant metastases in breast cancer patients: role of aromatase inhibitors. Cancer Treat Rev. 2007; 33:681–687.

100. Brown LF, Berse B, Van de Water L, Papadopoulos-Sergiou A, Perruzzi CA, Manseau EJ, et al. Expression and distribution of osteopontin in human tissues: widespread association with luminal epithelial surfaces. Mol Biol Cell. 1992; 3:1169–1180.

102. Chakraborty G, Jain S, Kundu GC. Osteopontin promotes vascular endothelial growth factor-dependent breast tumor growth and angiogenesis via autocrine and paracrine mechanisms. Cancer Res. 2008; 68:152–161.

103. Draffin JE, McFarlane S, Hill A, Johnston PG, Waugh DJ. CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Res. 2004; 64:5702–5711.

104. Tuck AB, Arsenault DM, O'Malley FP, Hota C, Ling MC, Wilson SM, et al. Osteopontin induces increased invasiveness and plasminogen activator expression of human mammary epithelial cells. Oncogene. 1999; 18:4237–4246.

105. Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003; 3:537–549.

106. Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011; 17:867–874.

107. Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009; 16:67–78.

108. Chen Q, Zhang XH, Massagué J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011; 20:538–549.

109. Wu K, Fukuda K, Xing F, Zhang Y, Sharma S, Liu Y, et al. Roles of the cyclooxygenase 2 matrix metalloproteinase 1 pathway in brain metastasis of breast cancer. J Biol Chem. 2015; 290:9842–9854.

110. Cojoc M, Mäbert K, Muders MH, Dubrovska A. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Semin Cancer Biol. 2015; 31:16–27.

111. Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010; 40:280–293.

112. Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. 2014; 20:460–473.

113. McCarthy N. Autophagy: directed development. Nat Rev Cancer. 2014; 14:74–75.
114. Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001; 61:439–444.
115. Yao KC, Komata T, Kondo Y, Kanzawa T, Kondo S, Germano IM. Molecular response of human glioblastoma multiforme cells to ionizing radiation: cell cycle arrest, modulation of the expression of cyclindependent kinase inhibitors, and autophagy. J Neurosurg. 2003; 98:378–384.

116. Lu J, Sun D, Gao S, Gao Y, Ye J, Liu P. Cyclovirobuxine D induces autophagy-associated cell death via the Akt/mTOR pathway in MCF-7 human breast cancer cells. J Pharmacol Sci. 2014; 125:74–82.

117. Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007; 21:1621–1635.

118. Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007; 21:1367–1381.

119. Ito H, Daido S, Kanzawa T, Kondo S, Kondo Y. Radiation-induced autophagy is associated with LC3 and its inhibition sensitizes malignant glioma cells. Int J Oncol. 2005; 26:1401–1410.

120. Chaterjee M, van Golen KL. Breast cancer stem cells survive periods of farnesyl-transferase inhibitor-induced dormancy by undergoing autophagy. Bone Marrow Res. 2011; 2011:362938.

121. Rao R, Balusu R, Fiskus W, Mudunuru U, Venkannagari S, Chauhan L, et al. Combination of pan-histone deacetylase inhibitor and autophagy inhibitor exerts superior efficacy against triple-negative human breast cancer cells. Mol Cancer Ther. 2012; 11:973–983.

122. Gong C, Bauvy C, Tonelli G, Yue W, Deloménie C, Nicolas V, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013; 32:2261–2272.

123. Palacios F, Tushir JS, Fujita Y, D'Souza-Schorey C. Lysosomal targeting of E-cadherin: a unique mechanism for the down-regulation of cell-cell adhesion during epithelial to mesenchymal transitions. Mol Cell Biol. 2005; 25:389–402.

124. Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007; 450:908–912.

125. van Vlerken LE, Hurt EM, Hollingsworth RE. The role of epigenetic regulation in stem cell and cancer biology. J Mol Med (Berl). 2012; 90:791–801.

126. Jin B, Ernst J, Tiedemann RL, Xu H, Sureshchandra S, Kellis M, et al. Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. Cell Rep. 2012; 2:1411–1424.

127. El Helou R, Wicinski J, Guille A, Adélaïde J, Finetti P, Bertucci F, et al. Brief reports: a distinct DNA methylation signature defines breast cancer stem cells and predicts cancer outcome. Stem Cells. 2014; 32:3031–3036.

128. Shukla S, Meeran SM. Epigenetics of cancer stem cells: pathways and therapeutics. Biochim Biophys Acta. 2014; 1840:3494–3502.

129. Han M, Liu M, Wang Y, Chen X, Xu J, Sun Y, et al. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One. 2012; 7:e39520.

130. Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009; 138:592–603.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download