Abstract
Purpose
Circular RNAs (circRNAs), a novel class of RNAs, perform important functions in biological processes. However, the role of circRNAs in the mammary gland remains unknown. The present study is aimed at identifying and characterizing the circRNAs expressed in the mammary gland of lactating rats.
Methods
Deep sequencing of RNase R-enriched rat lactating mammary gland samples was performed and circRNAs were predicted using a previously reported computational pipeline. Gene ontology terms of circRNA-producing genes were also analyzed.
Results
A total of 6,824 and 4,523 circRNAs were identified from rat mammary glands at two different lactation stages. Numerous circRNAs were specifically expressed at different lactation stages, and only 1,314 circRNAs were detected at both lactation stages. The majority of the candidate circRNAs map to noncoding intronic and intergenic regions. The results demonstrate a circular preference or specificity of some genes. DAVID analysis revealed an enrichment of protein kinases and related proteins among the set of genes encoding circRNAs. Interestingly, four protein-coding genes (Rev3l, IGSF11, MAML2, and LPP) that also transcribe high levels of circRNAs have been reported to be involved in cancer.
RNA functions as an intermediate molecule in the transfer of genetic information from DNA to protein. Noncoding RNAs (ncRNAs) are defined as RNAs that do not code proteins; they include all traditional classes of RNAs (except for mRNAs) such as rRNAs, tRNAs, and snRNAs, as well as the more recently discovered microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). With the advent of robust high throughput sequencing technologies and advances in bioinformatics, thousands of long and short ncRNAs have been discovered in the past decade in prokaryotes, eukaryotes, and viruses. These ncRNAs encompass transcripts of different sizes, from processed small RNAs ~20-30 bases in length to lncRNAs that are hundreds or even thousands of bases long [1]. These recently discovered ncRNAs represent a new paradigm of gene regulation. In particular, many miRNAs and lncRNAs have been reported to regulate both normal development and tumorigenesis in the mammary gland [2].
Circular RNA (circRNA) is a recently discovered ncRNA. CircRNA isoforms have largely gone unnoticed since their discovery in 1979, with some notable exceptions [3]. Recent studies by two independent teams demonstrated that a human circRNA running antisense to the cerebellar degenerationrelated protein 1 (CDR1) locus functions as a sponge for miR-7 [45]. Interestingly, miR-7 inhibits the epithelial-mesenchymal transition and metastasis of breast cancer, and sensitizes cells to DNA damage [67], which indicates that circRNAs may play profound roles in breast cancer. A circRNA transcribed from the Sry gene was also reported to act as a miR-138 sponge [8]. These two examples represent a strong paradigm for circRNA function, and have therefore attracted attention towards circRNAs. Growing evidence indicates that circRNAs, which are expressed from a large fraction of human, mouse, zebrafish, Caenorhabditis elegans, and fruit fly genes, constitute the major RNA isoform from these genes [9101112]. However, no information on mammary gland circRNAs has been reported.
Breastfeeding is not only beneficial to infants, but also provides many health benefits to mothers [13]. The maternal health benefits include decreased postpartum bleeding, more rapid uterine involution, decreased menstrual blood loss, increased child spacing, earlier return to prepregnancy weight, and possible decreased risk of hip fractures and osteoporosis in the postmenopausal period [13]. One of the most notable maternal health benefits is a decreased risk of breast cancer, which has been demonstrated by several groups using different methods [14]. These findings suggest that breastfeeding affects biological processes in the mammary gland. However, the molecular mechanisms underlying the role of breastfeeding in breast cancer prevention remain unclear. Accordingly, an understanding of the molecular landscape of the normal breast during lactation is critical for breast cancer prevention.
In the present study, we investigate the circRNA expression pattern in the lactating mammary gland to provide a basis for comparison between breast cancer profiles, and for selection of representative circRNA candidates for future functional characterization in breast development and breast cancer.
Mammary gland tissue samples from ten Sprague Dawley rats (Rattus norvegicus) fifth parturition were obtained from Vital River Laboratories (Beijing, China). Lactating mammary glands were harvested from five rats at day 1 and 7 postpartum and frozen in liquid nitrogen for subsequent extraction of RNA. All samples were obtained in accordance with the experimental procedures guideline of Jiangsu Normal University, and all experimental protocols were approved by the Animal Ethics Committee of Jiangsu Normal University.
Total RNA was extracted using Trizol (Life Technologies, Carlsbad, USA), and treated with DNase I. The RNase R enrichment protocol was modified from a previous report [11]. Each total RNA sample (20 µg) was depleted for ribosomal RNAs using the RiboMinus kit (Life Technologies), and then subjected to RNase R treatment. RiboMinus RNA was incubated for 15 minutes at 37℃, with or without (control) 3 U/µg of RNase R (Epicentre Biotechnologies). RNA was subsequently purified by phenol-chloroform extraction, and divided into two parts, one of which was used to prepare libraries for sequencing, using Illumina's whole-transcriptome RNA sequencing protocols (Illumina, San Diego, USA). The Illumina Hiseq2000 System was used to perform sequencing runs for RNA sequencing. The other was reverse transcribed using the SuperScript III FirstStrand Synthesis System (Life Technologies) with random hexamers, according to the manufacturer's instructions.
The rat reference genome was downloaded from the National Center for Biotechnology Information (NCBI) genome browser (ftp://ftp.ncbi.nlm.nih.gov/genomes/R_norvegicus/). Sequencing reads from each pooled sample were mapped independently using the short-read mapper Bowtie 2 [15]. Reads aligning contiguously and full length to the genome were discarded. We extracted 20-mers from both ends and aligned them independently to find unique anchor positions within spliced exons from the remaining reads. Anchors that aligned in the reverse orientation (head-to-tail) indicated circRNA splicing. We extended the anchor alignments such that the complete read aligns and the breakpoints were flanked by GU/AG splice sites. Ambiguous breakpoints were discarded. More information can be found in a previous report [5].
The DAVID tool (National Institute of Allergy and Infectious Diseases, Bethesda, USA) was used for enrichment of gene ontology (GO) terms of circRNA-producing genes.
FASTA files of rat miRNAs were obtained from miRBase release 20.0 (http://www.mirbase.org/). Only mature miRNAs were considered for seed analysis. The miRNAs were aligned with circRNAs. A putative target site of a miRNA is a 6-nucleotide-long sequence in the genome that represents the reverse complement of nucleotides 2-7 of the mature miRNA sequence.
In order to test the global relationship between the circRNAs and linear RNAs, we fit a Poisson model per gene modeling circular expression by reads per kilobase of exon model per million mapped reads (RPKM) to poly(A) gene expression using SPSS version 19.0 (IBM Corp., Armonk, USA).
cDNAs of mock and RNase R-treated RNA were diluted with water and used as a template for polymerase chain reaction (PCR). In order to validate the unpolyadenylated status of circRNAs, the total RNAs were reverse-transcribed with oligo-dTs and random hexamer primers, and the cDNAs were subjected to qPCR. Standard PCR was performed using Taq DNA polymerase (New England Biolabs, Beverly, USA), and quantitative PCR was performed using SYBR Green Mix (Roche, Basel, Switzerland) on a StepOne plus Real-Time PCR System. qPCR Ct values were calculated automatically using the manufacturer's software. The PCR primers are listed in Supplementary Table 1 (available online). PCR products were directly Sanger-sequenced in both directions using amplification primers.
We obtained 11,853,653 and 11,970,307 clean sequence reads of 86 nucleotides from the two RNase R-treated RiboMinus mammary gland RNA pools. Nearly half (45%) of the clean reads were predicted as circRNAs, and most of the circRNAs were transcribed from gene-containing regions covering 1,569 genes. These data were consistent with findings from previous studies that identified circRNAs from mainly exons and introns of gene-containing regions (Figure 1) [34591011].
We detected 6,824 and 4,523 circRNAs from lactating mammary glands at day 1 (D1) and 7 (D7) postpartum, respectively. The length of circRNAs varied from 66 nt to 1,955 nt. Numerous circRNAs seemed to be specifically expressed at different lactation stages, as only 1,314 circRNAs were detected at both lactation stages.
The circularization of circRNAs and their relative expression levels, as obtained from RNA-sequencing, were validated. Inward and outward facing primers were designed against 11 randomly selected representative transcripts. Each primer pair amplified a single, distinct product of the expected size and sequence. RNase R resistance of all 11 novel backsplice events was apparent following RNase R treatment, whereas the abundance of linear RNAs was found to decrease (Table 1). Moreover, oligo-dT priming for reverse transcription resulted in significantly lower levels of backspliced products relative to poly(A)-containing transcript levels, indicating that these species were not polyadenylated (Table 1). The difference in the expression of the 11 circRNAs between the two time points was also confirmed by qPCR (Supplementary Figure 1).
In order to exclude the possibility that circRNAs might be the result of the background "noise" level of dysfunctional splicing, we analyzed the relationship between linear RNA isoform expression from a given gene and the probability of detecting a circRNA isoform from that gene. We did not find any correlation between the expressions of the two RNA isoforms (Figure 2).
In order to investigate the function of circRNAs, the GO term enrichment of the set circRNA-producing genes was analyzed using the DAVID tool. The top five annotation clusters, summarized in Table 2, involved nucleotide and ATP binding, nuclear lumen, GTPase regulator activity, ion binding and protein kinase activity.
In the present study, 716 out of 728 mature rat miRNAs were predicted to have docking sites in the circRNAs detected in the rat mammary gland. Only a few circRNAs were predicted to have many miRNA-binding sites, which indicated that circularization was not a global feature under selection in the sequence of the thousands of circRNAs profiled.
Previous studies have identified circRNAs mainly from exons and introns in gene-containing regions [34591011]. In the present study, circRNAs were not only identified from gene containing regions, but also from intergenic regions. The circRNAs distributed in noncoding regions accounted for ~70% of the total circRNAs, including those from intronic and intergenic regions, of which intronic circRNAs accounted for more than 40%. This is similar to the expression pattern of other ncRNAs [16].
The top five annotation clusters involved nucleotide and ATP binding, nuclear lumen, GTPase regulator activity, ion binding and protein kinase activity. As the activation of virtually all cell-surface receptors leads directly or indirectly to changes in protein phosphorylation via activation of protein kinases, the circRNAs derived from protein kinase-encoding genes may also be functional [17]. We also analyzed mouse and human circRNA data from several tissues [51011]. Notably, DAVID analysis revealed an enrichment of protein kinases and related proteins among the set of genes producing circRNAs in the tissues of various species. This finding further verified that circRNAs show a high degree of evolutionary conservation, suggesting that they might have important biological functions.
In the present study, most circRNAs were transcribed from protein-coding gene regions, which indicated a circular preference or specificity of some genes. The set of genes producing circRNAs in the tissues of various mammal species were all enriched for protein kinases and related proteins [511]. As shown in Table 3, seven of the 10 most highly expressed circRNA families were transcribed from seven different protein-coding gene regions. Interestingly, four of the protein-coding genes (Rev3l, IGSF11, MAML2, and LPP) were reported to be involved in carcinogenesis. Rev3l, which encodes an error-prone DNA polymerase Pol ζ subunit, is critical for embryonic development and maintenance of genome stability [1819]. Loss of Rev3l enhances spontaneous mammary tumorigenesis, and genetic variation in the Rev3l gene is reported to affect the development and progression of breast cancer [2021]. The expression of the immunoglobulin superfamily 11 gene (IGSF11) was elevated in colorectal cancers and hepatocellular carcinomas as well as in intestinal-type gastric cancers, and IGSF11 is suggested to be a potential target for cancer immunotherapy [22]. MAML2, a member of the mastermind-like (MAML) family, functions as an essential coactivator of NOTCH in regulating a variety of malignancies [23]. There is increasing evidence that lipoma preferred partner (LPP) plays an important role in the development of a variety of human cancers [24]. A recent report revealed that LPP, which was normally operative in cells of mesenchymal origin, could be co-opted by breast cancer cells during an epithelial-mesenchymal transition to promote their migration and invasion [25]. Expression levels of the circRNA families transcribed from the above-mentioned four genes were all remarkably higher than those in their linear counterparts. The abundance of circRNA family from Igsf11 was 1,700-fold higher than that of Igsf11 mRNA expressed in the mammary gland. These data indicate that although the functions of these circRNAs remain unclear, they are unlikely to be the result of background "noise."
Recent reports have shown that, in mammals, circRNAs may function as miRNA sponges or compete against endogenous RNAs [456]. However, the enrichment of miRNA binding sites was not a global feature under selection in the sequence of the thousands of circRNAs profiled in the present study, which is consistent with the findings of a recent report [10]. However, we did identify a highly expressed circRNA that had several miRNA-binding sites. CircRNA_1093, with a short length of 108 bp, had 4 miR-342-3p binding sites. Notably, miR-342 was recently reported to regulate BRCA1 expression in breast cancer [26]. Therefore, we speculate that these circRNAs may be involved in the regulation of oncogenesis. In addition to their general regulatory functions in normal development, miR-200 and let-7 families are relatively well characterized in breast cancer initiation and self-renewal [2728]. A total of 3,100 circRNAs possess putative binding sites for members of the miR-200 family, of which 599 circRNAs have more than 10 binding sites, and a total of 2,539 circRNAs have putative binding sites for members of the let-7 family, of which 932 circRNAs have more than 10 binding sites. The high stability of circRNA in cells [11] suggests that they might be involved in the regulation of breast cancer via miRNAs. However, the potential genome-wide interplay between miRNAs and circRNAs needs further experimental and computational investigation.
Although the function of circRNAs remains largely unclear, an association between circRNAs and cancers has been reported. There is a global reduction in circRNA abundance in colorectal cancer cell lines and cancer compared with that in normal tissues, revealing a negative correlation between global circRNA abundance and proliferation and cancer [29]. Additionally, typical circRNAs were found to be show a significantly lower expression in gastric cancer tissues than in the paired adjacent nontumorous tissues [30]. These findings indicate that circRNA is a promising biomarker for cancer diagnosis. In the present study, a total of 6,824 and 4,523 circRNAs were identified by deep RNA sequencing from normal rat mammary glands at two lactation stages. These results provide the basis for comparison of breast cancer profiles and for selection of representative circRNA candidates for future functional characterization in breast development and breast cancer.
Figures and Tables
Figure 1
The distribution of circular RNAs in the genome. (A) Rat mammary gland library at day 1 postpartum. (B) Rat mammary gland library at day 7 postpartum.
CDS=coding sequences.
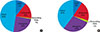
Figure 2
Quantitative regulation of circular to linear isoform. No genome-wide trend between circular expression and linear transcript expression as measured by log PolyA reads per kilobase of exon model per million mapped reads (RPKM).
Y axis=RPKM for circular RNA; X axis=log PolyA RPKM of linear isoform.

Table 1
Quantitative polymerase chain reaction validation of circular RNAs by different reverse transcription primers and RNase R treatment

Table 2
Gene ontology term enrichment of circular RNA generating genes

Table 3
The 10 most abundance co-expressed circular RNA families in rat mammary gland at day 1 and day 7

Notes
This work was partly funded by National Natural Science Foundation of China (31472077, 31101703), Natural Science Foundation of Jiangsu Province (BK2011206), Jiangsu Overseas Research and Training Program for University Prominent Young and Middle-aged Teachers and Presidents Project, and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
1. Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol. 2014; 49:16–32.

2. Piao HL, Ma L. Non-coding RNAs as regulators of mammary development and breast cancer. J Mammary Gland Biol Neoplasia. 2012; 17:33–42.

3. Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979; 280:339–340.

4. Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011; 30:4414–4422.

5. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338.

6. Kong X, Li G, Yuan Y, He Y, Wu X, Zhang W, et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS One. 2012; 7:e41523.

7. Yu N, Huangyang P, Yang X, Han X, Yan R, Jia H, et al. microRNA-7 suppresses the invasive potential of breast cancer cells and sensitizes cells to DNA damages by targeting histone methyltransferase SET8. J Biol Chem. 2013; 288:19633–19642.

8. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388.

9. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012; 7:e30733.

10. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013; 9:e1003777.

11. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013; 19:141–157.

12. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014; 15:409.

13. Gartner LM, Morton J, Lawrence RA, Naylor AJ, O'Hare D, Schanler RJ, et al. Breastfeeding and the use of human milk. Pediatrics. 2005; 115:496–506.

14. Kotsopoulos J, Lubinski J, Salmena L, Lynch HT, Kim-Sing C, Foulkes WD, et al. Breastfeeding and the risk of breast cancer in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. 2012; 14:R42.

15. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012; 9:357–359.

16. St Laurent G, Shtokalo D, Tackett MR, Yang Z, Eremina T, Wahlestedt C, et al. Intronic RNAs constitute the major fraction of the non-coding RNA in mammalian cells. BMC Genomics. 2012; 13:504.

17. Clark DP. Molecular Biology: Academic Cell Update. Burlington: Academic Press/Elsevier;2010.
18. Schenten D, Kracker S, Esposito G, Franco S, Klein U, Murphy M, et al. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J Exp Med. 2009; 206:477–490.

19. Lange SS, Bedford E, Reh S, Wittschieben JP, Carbajal S, Kusewitt DF, et al. Dual role for mammalian DNA polymerase zeta in maintaining genome stability and proliferative responses. Proc Natl Acad Sci U S A. 2013; 110:E687–E696.
20. Wittschieben JP, Patil V, Glushets V, Robinson LJ, Kusewitt DF, Wood RD. Loss of DNA polymerase zeta enhances spontaneous tumorigenesis. Cancer Res. 2010; 70:2770–2778.

21. Varadi V, Bevier M, Grzybowska E, Johansson R, Enquist K, Henriksson R, et al. Genetic variation in genes encoding for polymerase zeta subunits associates with breast cancer risk, tumour characteristics and survival. Breast Cancer Res Treat. 2011; 129:235–245.

22. Watanabe T, Suda T, Tsunoda T, Uchida N, Ura K, Kato T, et al. Identification of immunoglobulin superfamily 11 (IGSF11) as a novel target for cancer immunotherapy of gastrointestinal and hepatocellular carcinomas. Cancer Sci. 2005; 96:498–506.

23. Köchert K, Ullrich K, Kreher S, Aster JC, Kitagawa M, Jöhrens K, et al. High-level expression of Mastermind-like 2 contributes to aberrant activation of the NOTCH signaling pathway in human lymphomas. Oncogene. 2011; 30:1831–1840.

24. Grunewald TG, Pasedag SM, Butt E. Cell adhesion and transcriptional activity: defining the role of the novel protooncogene LPP. Transl Oncol. 2009; 2:107–116.

25. Ngan E, Northey JJ, Brown CM, Ursini-Siegel J, Siegel PM. A complex containing LPP and alpha-actinin mediates TGFbeta-induced migration and invasion of ErbB2-expressing breast cancer cells. J Cell Sci. 2013; 126(Pt 9):1981–1991.
26. Crippa E, Lusa L, De Cecco L, Marchesi E, Calin GA, Radice P, et al. miR-342 regulates BRCA1 expression through modulation of ID4 in breast cancer. PLoS One. 2014; 9:e87039.

27. Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008; 283:14910–14914.

28. Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007; 131:1109–1123.

29. Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, et al. Correlation of circular RNA abundance with proliferation: exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep. 2015; 5:8057.
Supplementary Material
Supplementary Figure 1
Correlation of relative expression value quantified by qPCR with that analyzed by RNA sequencing. Y-axis, representing the circRNA abundance differentiation between rat mammary glands at day 1 and day 7 as measured by qPCR; X-axis, representing the circRNA abundance differentiation between rat mammary glands at day 1 and day 7 as measured by RNA-sequencing.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download