This article has been corrected. See "Erratum: Lentivirus-Mediated Short-Hairpin RNA Targeting Protein Phosphatase 4 Regulatory Subunit 1 Inhibits Growth in Breast Cancer" in Volume 19 on page 467.
This article has been retracted. See "Retraction note to: Lentivirus-Mediated Short-Hairpin RNA Targeting Protein Phosphatase 4 Regulatory Subunit 1 Inhibits Growth in Breast Cancer" in Volume 21 on page 102.
Abstract
Purpose
Protein phosphatase 4 regulatory subunit 1 (PP4R1), as an interaction partner of the catalytic serine/threonine-protein phosphatase 4 catalytic subunit has been shown to involve in cellular processes and nuclear factor κB signaling. However, the functions of PP4R1 in human breast cancers remain unclear. This study is designed to explore the effect of PP4R1 knockdown on the biological characteristics of breast cancer cells.
Methods
A lentivirus-mediated short hairpin RNA (shRNA) was designed to knockdown the expression of PP4R1 in ZR-75-30 breast cancer cells. The efficiency of lentivirus-mediated shRNA infection was determined using fluorescence microscopy to observe lentivirus-mediated green fluorescent protein expression and confirmed to be over 80%. PP4R1 expression in infected ZR-75-30 cells was detected by quantitative real-time polymerase chain reaction and western blot analysis. Cell proliferation and colony formation ability were measured by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) assay and colony formation assay, respectively. Flow cytometry was used to measure cell cycle progression and cell apoptosis. In addition, apoptosis makers, including poly-ADP-ribose polymerase (PARP) and caspase-3, were investigated in PP4R1-silenced ZR-75-30 cells by western blot assay.
Results
We successfully constructed lentivirus-mediated shRNA to target PP4R1 in ZR-75-30 cells. MTT assay and colony formation assay showed the loss of PP4R1 suppressed the proliferation of ZR-75-30 cells. Flow cytometry analysis indicated cell cycle arrest and increased cell apoptosis in PP4R1 knockdown cells. Further, the apoptosis response in cells depleted of PP4R1 was illustrated by downregulation of PARP and upregulation of caspase-3.
Breast cancer is a leading cause of female mortality and is considered as the most frequent life-threatening cancer in women [1]. Every year, breast cancer results in about 10% female cancer deaths [23]. However, there is no effective therapy against breast cancer. Molecular-target therapy, has now emerged as a novel concept [4], and is a useful tool in the discovery of anticancer agents.
Previous reports showed that serine/threonine protein phosphatase 4 (PP4), a holoenzyme composed of a catalytic subunit PP4C, and other regulatory subunits, plays an important role in the development of prostate carcinoma and leukemia [56]. PP4C is an essential polypeptide, which regulate critical cellular processes including apoptosis and DNA damage checkpoint recovery [7]. Wang et al. [8] reported that PP4C is overexpressed in breast and lung cancer, and its inhibition sensitized breast and lung cancer cells to cisplatin treatment, suggesting PP4C overexpression may play an important role in human malignancies. Protein phosphatase 4 regulatory subunit 1 (PP4R1) derived from bovine testis extracts and a molecular weight of 125 kDa, was the first described stable interactor of PP4C [9]. In addition, the complex containing PP4, PP4C, and PP4R1 was reported to regulate histone deacetylase 3 activity [10] and affect microtubule growth of the centrosome [11]. Despite its importance, the molecular mechanisms by which PP4R1 contributes to breast cancer remain unclear. Gene silencing has been widely applied in clinical trials as a potential therapy for a number of diseases [1112]. Compared to chemical inhibitors, gene silencing has the advantages of long-term effect and specificity, and its efficiently regulates various isoforms of target genes [13]. RNA interference (RNAi) is a powerful tool for target validation in cell culture and is widely used in gene silencing experiment [14]. RNAi can be applied in mammalian cells using small interfering RNA (siRNA) and short hairpin RNA (shRNA) [151617]. The shRNA-mediated silencing has the benefits of achieving longer silencing, providing more delivery options and lower costs, compared to synthetic siRNA.
In this study, we constructed a lentivirus-mediated shRNA targeting PP4R1 to suppress PP4R1 expression in ZR-75-30 cells, resulting in reduced cell proliferation, arrested cell cycle, and increased cell apoptosis. We further explored its molecular mechanism in the cell cycle and apoptosis. Our study indicates that PP4R1 may serve as a potential target for gene therapy in treatment of breast cancer.
The human breast cancer cell line ZR-75-30, T-47D and the human embryonic kidney cell line 293T (HEK293) were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). RPMI-1640 (Hyclone, Logan, USA) containing 10% fetal bovine serum (FBS) was used to culture ZR-75-30 cells. Both T-47D and HEK293 cells were grown in DMEM (Hyclone) containing 10% FBS. All cells were maintained in a humidified atmosphere with 5% CO2 at 37℃.
The lentiviral backbone plasmid pFH-L, pHelper plasmids pVSVG-I and pCMVΔR8.92 were provided by Shanghai Hollybio Co., Ltd. (Shanghai, China). The pFH-L containing RNA polymerase III promoter H1, drives the shRNA insertion sequence expressing siRNA which can continuously play interference effects in host cells. Moreover, the pFH-L expresses the reporter gene green fluorescent protein (GFP) by the cytomegalovirus (CMV) promoter.
The human PP4R1-specific targeting sequence was designed using online siRNA tools with PP4R1 sequence (NM_001042388.2) as a reference. The sequence of the shRNA cassette against human PP4R1 (shPP4R1) is 5'-GCT TGA ATC TCG GTG TCT TTC CTC GAG GAA AGA CAC CGA GAT TCA AGC TTT TTT-3' and was ligated into pFH-L vector (Shanghai Hollybio Co., Ltd.) by NheI and PacI linearization. The negative control shRNA sequence (shCon) is 5'-GCG GAG GGT TTG AAA GAA TAT CTC GAG ATA TTC TTT CAA ACC CTC CGC TTT TTT-3'. The complementary oligonucleotides pairs of these sequences were ligated into the linearized pFH-L vector after synthesis and annealing. The pFH-L can express GFP, used as a gene reporter, under the CMV promoter, The constructed lentiviral plasmids were thereafter denoted as pFH-L-shPP4RL, were used for specific interference of PP4R1 expression and negative control, respectively. Lipofectamine 2000 (Invitrogen, Carlsbad, USA) was used to co-transfect pFH-L-shPP4R1 or pFH-L-shCon into 293T cells with pHelper plasmids pVSVG-I and pCMVΔR8.92 (Shanghai Hollybio Co., Ltd.). At 48-hour posttransfection, lentiviral particles were collected from the media and further purified by ultracentrifugation, and named as shPP4R1 and shCon. Viral titer was calculated by counting cells positive for GFP under fluorescent microscope based on serial dilutions method.
ZR-75-30 cells (5×104 cells/well) in the logarithmic growth phase were grown in 6-well plates overnight and infected with shPP4R1 at a multiplicity of infection of 35. Infection efficiency was determined by observing the percentage of GFP-positive cells under fluorescent microscope. Cells were harvested 96 hours after infection and the knockdown efficiency of PP4R1 was evaluated by western blot and quantitative realtime polymerase chain reaction (qRT-PCR) analysis.
After 4 days infection, Trizol reagent (Invitrogen) was used to extract total RNA from all cells. The procedure was carried out using the following primers: PP4R1, 5'-ACGTCC CATTGCTCTGAATC-3' (forward), and 5'-CTT GGGACATCT GCCAAAGT-3' (reverse). The β-actin was used as an internal reference and its primer sequences were 5'-GTG GAC ATC CGC AAA GAC-3' (forward) and 5'-AAAGGGTGTAACGCAACTA-3'. Total 25 µL reaction mixtures, containing 0.8 µL forward and reverse primers (2.5 µM), 5 µL cDNA, 10 µL 2×SYBR premix ex taq and 4.2 µL ddH2O, carried out the reaction conditions with initial 1 minute denaturation at 95℃ and followed by 40 cycles containing denaturation at 95℃ for 20 seconds and annealing extension at 60℃. The absorbance value was read at the extension stage and 2-ΔΔCt method was used to calculate the PP4R1 mRNA relative expression level.
Cells were scraped on ice, collected by centrifugation and lysed in 2×SDS sample buffer containing 4% SDS, 10% glycerol, 10 mM EDTA, and 100 mM Tris-HCl, pH 6.8 and collected by centrifugation. Subsequently, protein concentrations were determined by protein assay kit and total 30 µg proteins were separated on 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes at 300 mA for 2 hours. The blots were probed with primary antibodies overnight at 4℃ after being blocking in Tris buffered saline with Tween and 5% skim milk. Then the membrane was incubated with secondary antibody for 1 hour at room temperature. The primary antibodies were rabbit anti-PP4R1 (#HPA041089, 1:1,000; Sigma, St. Louis, USA), rabbit anti-caspase-3 (#9661, 1:500; Cell Signaling, Danvers, USA), rabbit anti-PARP (#9542, 1:1,000; Cell Signaling) and rabbit anti-GAPDH (#10494-1-AP, 1:500,000; Proteintech Group Inc., Chicago, USA) and horseradish peroxidase-conjugated goat anti-rabbit IgG (#SC-2054, 1:5,000; Santa Cruz Biotechnology, Santa Cruz, USA) was used as second antibody. The protein expression level was detected by enhanced chemiluminescence plus TM blotting system (Little Chalfont, Bucks, UK).
ZR-75-30 cells were cultured in 96-well plates containing fresh media at a density of 2×103 cells/well after 96 hours infection and used for 3-[4, 5-dimethylthiazol-2-yl]-2, 5-diphenyl tetrazolium bromide (MTT) assay. Briefly, each well was added 20 µL MTT (5 mg/mL) (Sigma) and incubated for 4 hours at 37℃, followed by adding 100 µL acidic isopropanol (5% isopropanol, 10% SDS, and 0.01 mol/L HCl). The absorbance of each well at 595 nm was measured by the Microplate Reader (Epoch, Winooski, USA).
The colony formation assay for the ZR-75-30 cells was performed as follows: after 96 hours of infection, cells (400 cells/well) were grown in 6-well plates for 6 days to form natural colonies. Then, colonies were fixed with 4% paraformaldehyde and stained with crystal violet. Colonies on the plate were counted under a microscope. Each colony consisted of more than 50 cells.
The infected ZR-75-30 cells (2×105 cells/dish) were seeded in 6 cm dishes for 40 hours and then were harvested and washed with PBS for three times, fixed in 70% ice-cold ethanol for 1 hour at 4℃. After being washed with PBS, cells were resuspended in 1 mL PBS containing 500 U/mL RNase. Cellular DNA was stained with of propidium iodide (PI) (20 µg/mL) for 30 minutes at room temperature after 30 minuntes incubation at 37℃. DNA content was determined using the CellQuest software on a FACS flow cytometer (BD Biosciences, San Jose, USA).
The infected ZR-75-30 cells were seeded in 6 cm dishes at a density of 3×104 cells/dish for 5 days. Annexin V-APC/7-AAD Apoptosis Detection Kit (#KGA1026; KeyGEN Biotech, Nanjing, China) was used to determine the rate of cellular apoptosis. In brief, cells were washed with PBS for three times and then suspended in 0.5 mL annexin binding buffer, 5 µL annexin V-APC solution and 5 µL 7-ADD solutions successively. Subsequently, cells were incubated in the medium for 15 minutes at room temperature in the dark and analyzed by flow cytometry (Caliber; Becton Dickinson, San Jose, USA).
Statistical analysis was performed using SPSS software version 16.0 (SPSS Inc., Chicago, USA). All data were expressed as the mean±standard deviation from three independent experiments. Student t-test was used to evaluate statistically significant differences between groups, and p<0.05 was considered statistically significant.
Real-time PCR analysis showed that ZR-75-30 cells demonstrated higher PP4R1 mRNA expression compared with T-47D cells (p<0.010) (Figure 1A). Also we found there was a higher protein expression in ZR-75-30 cells, compared with T-47D cells (Figure 1B), similar to results with PCR analysis. Therefore, we chose ZR-75-30 cells as the target cell for the below RNAi research.
The lentiviral shRNA was successfully constructed and transduced into ZR-75-30 cells (Figure 1C). To evaluate the silencing efficiency, PP4R1 mRNA and protein expressions were analyzed by qRT-PCR and western blot. As shown in Figure 1D and 1E, both the mRNA level and protein expression of PP4R1 were significantly decreased in the shPP4R1 group compared to the shCon group (p<0.001).
To examine the effect of downregulation of PP4R1 on the proliferation of breast cancer cells, ZR-75-30 cells were plated for MTT and colony formation assays after 96 hours infection. The results showed that the cell viability in shPP4R1 group started to reduce on day 3 with significant suppression on day 5 (p<0.001) (Figure 2A). Similarly, the colony formation rate of the shPP4R1 group was dramatically suppressed compared to the shCon group (Figure 2B). The number of colonies in the shPP4R1 group was much fewer than that in the shCon group (p<0.001) (Figure 2C). Collectively, these results showed that knockdown of PP4R1 by lentivirus-mediated RNAi could significantly inhibit breast cancer cells growth in vitro.
To explore the effect of knockdown of PP4R1 on cell cycle progression, we assessed the cell cycle distribution in three groups of ZR-75-30 cells (Figure 3A). As shown in Figure 3B, with the absence of PP4R1 in ZR-75-30 cells, the percentage of cells in G0/G1 phase was obviously expanded (p<0.010). Additionally, the shPP4R1 group exhibited higher proportion of sub-G1 phase (p<0.050) compared with the Con and shCon groups (Figure 3C), suggesting that PP4R1 silencing induced cell apoptosis.
To verify the role of PP4R1 silencing in cell apoptosis, flow cytometry analysis for Annexin V/PI double staining-coupled was performed in ZR-75-30 cells (Figure 4A). The apoptotic cells contained the early apoptotic cells (Annexin V+/PI-) and the late apoptotic cells (Annexin V+/PI+). As shown in Figure 4B, both the rates of early and late apoptotic cells in shPP4R1 group were significantly higher than those in the Con and shCon groups (p<0.001). Our results suggested that PP4R1 silencing remarkably promoted breast cancer cell apoptosis.
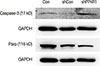
Furthermore, we analyzed the expression of apoptotic markers, as shown in Figure 5. The expression of caspase-3 was upregulated and the cleaved PARP was downregulated in shPP4R1 group compared with shCon group in ZR-75-30 cells.
Breast cancer is the most frequently diagnosed life-threatening cancer in women. Although breast tumors initially respond to chemotherapy, incomplete pathologic response leads to higher tumor recurrence [1819]. To date, there are no effective biomarkers for breast cancer prognosis.
PP4R1, a 125-kDa protein that co-purifies with PP4C from bovine testis extracts has been reported to be linked to microtubule growth [920]. To investigate its functional role in breast cancer, lentivirus-mediated shRNA targeting for PP4R1 was performed to knockdown its expression in the breast cancer cell line ZR-75-30. MTT and colony formation assay suggested that PP4R1 was indispensable for cell proliferation. Then, we examined cell cycle and apoptosis which played an important role in assessing the cellular metabolism, physiology and pathology. We found that downregulation of PP4R1 increased the percentage of G0/G1 phase cells, suggesting that PP4R1 might affect DNA synthesis. This might be explained by the previous report that the biological function of PP4R1 was mainly linked with repair of DNA double-strand breaks [21]. Besides, there was a higher proportion of sub-G1 phase cells in the cell cycle of PP4R1 silenced cells, revealing that deficiency for PP4R1 can cause cell apoptosis. The presence of cleaved PARP, as one of the most used diagnostic tools for detection of apoptosis, has been demonstrated to be upregulated in breast cancer [22]. Likewise, we discovered PARP was downregulated in PP4R1 silenced cells, which may be consistent with upregulation of cleaved PARP expression. It is generally accepted that cancer does not necessarily arise as a result of cell proliferation increase. Rather, there is an essential balance between the rate of cell-cycle progression and cell apoptosis [23]. To examine the cell apoptosis of PP4R1 knockdown cells, Annexin V/PI double staining-coupled flow cytometry analysis was performed and we found that PP4R1 silencing could promote cell apoptosis. Subsequently, caspase-3, an apoptosis maker, was detected by the western blot analysis. The expression level of caspase-3 was significantly upregulated in PP4R1 silencing breast cancer cells. Which was consistent with related study showing its activation is a crucial event leading to tumor cell apoptosis [24]. Our results further confirmed that the suppression of cell proliferation in PP4R1 silencing cells might be caused by increased apoptotic cell death by upregulating caspase-3.
In summary, this is the first report of an shRNA-mediated PP4R1 silencing strategy to inhibit cell growth and survival in ZR-75-30 breast cancer cells, which may be used as a new anticancer molecular therapy. Further validation of the effect of PP4R1 in other breast cancer cells needs to be conducted.
Figures and Tables
Figure 1
Establishment of protein phosphatase 4 regulatory subunit 1 (PP4R1)-silenced breast cancer cells. (A) Quantitative real-time polymerase chain reaction (qRT-PCR) analysis of PP4R1 was performed in T-47D and ZR-75-30 cells, respectively. (B) Western blot analysis of PP4R1 was performed in T-47D and ZR-75-30 cells, respectively. (C) Fluorescence microscopy examination was performed for the infection efficiency lentivirus vector in ZR-75-30 cells (scale bar, 10 µm). (D) qRT-PCR analysis was performed after the knockdown of PP4R1 by lentivirus-mediated RNA interference (RNAi) in ZR-75-30 cells. (E) Western blot analysis of the knockdown of PP4R1 was performed by lentivirus-mediated RNAi in ZR-75-30 cells.
GAPDH=glyceraldehyde-3-phosphate dehydrogenase; Con=control; sh=short hairpin; shCon=the lentivirus-mediated negative control RNAi group; shPP4R1=the lentivirus-mediated PP4R1 RNAi group; GFP=green fluorescent protein.
*p<0.01; †p<0.001, compared with shCon.
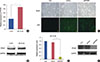
Figure 2
Suppression of proliferation and colony formation of ZR-75-30 cells by protein phosphatase 4 regulatory subunit (PP4R1) silencing. (A) Proliferation of PP4R1-silenced ZR-75-30 cells was assessed by MTT assay, in parallel with Con and shCon group as control. (B) The image of the colony formation assay is represented (scale bar, 50 µm; magnification, ×40). (C) Statistical analyses of colonies number results in ZR-75-30 cells, in parallel with Con and shCon group as control, is represented.
Con=control; sh=short hairpin; shCon=the lentivirus-mediated negative control RNAi group; shPP4R1=the lentivirus-mediated PP4R1 RNAi group.
*p<0.001, compared with shCon.
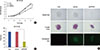
Figure 3
Cell cycle arrest of ZR-75-30 cells induced by protein phosphatase 4 regulatory subunit 1 (PP4R1) silencing. (A) Flow cytometry analysis of cell cycle progression was performed on the three groups (Con, shCon, and shPP4R1 groups). (B, C) Statistical analysis of cell cycle including the apoptotic cells (sub-G1 cell) is represented.
Con=control; sh=short hairpin; shCon=the lentivirus-mediated negative control RNAi group; shPP4R1=the lentivirus-mediated PP4R1 RNAi group.
*p<0.05, compared with shCon.
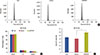
Figure 4
Apoptosis in ZR-75-30 cells induced by protein phosphatase 4 regulatory subunit 1 (PP4R1) silencing. (A) Representative flow cytometric dot plots are presented. Cell in the upper right and lower right quadrant were characterized as early apoptotic cells and late apoptotic cells, respectively. (B) Statistical analysis on early and late apoptotic cell percentage in ZR-75-30 cells is represented.
Con=control; sh=short hairpin; shCon=the lentivirus-mediated negative control RNAi group; shPP4R1=the lentivirus-mediated PP4R1 RNAi group.
*p<0.001.

Figure 5
Western blot analysis of several critical apoptosis markers, including caspase-3 and poly-ADP-ribose polymerase (PARP) with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the internal control. The figure shows the representative images of three independent experiments.
Con=control; sh=short hairpin; shCon=the lentivirus-mediated negative control RNAi group; shPP4R1=the lentivirus-mediated PP4R1 RNAi group.
References
1. Li CI, Daling JR, Malone KE. Age-specific incidence rates of in situ breast carcinomas by histologic type, 1980 to 2001. Cancer Epidemiol Biomarkers Prev. 2005; 14:1008–1011.

2. Anderson BO, Yip CH, Ramsey SD, Bengoa R, Braun S, Fitch M, et al. Breast cancer in limited-resource countries: health care systems and public policy. Breast J. 2006; 12:Suppl 1. S54–S69.

3. Parkin DM, Fernández LM. Use of statistics to assess the global burden of breast cancer. Breast J. 2006; 12:Suppl 1. S70–S80.

4. Allgayer H, Fulda S. An introduction to molecular targeted therapy of cancer. Adv Med Sci. 2008; 53:130–138.

5. Inostroza J, Sáenz L, Calaf G, Cabello G, Parra E. Role of the phosphatase PP4 in the activation of JNK-1 in prostate carcinoma cell lines PC-3 and LNCaP resulting in increased AP-1 and EGR-1 activity. Biol Res. 2005; 38:163–178.

6. Mourtada-Maarabouni M, Williams GT. Protein phosphatase 4 regulatesapoptosis in leukemic and primary human T-cells. Leuk Res. 2009; 33:1539–1551.

7. Chen GI, Tisayakorn S, Jorgensen C, D'Ambrosio LM, Goudreault M, Gingras AC. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J Biol Chem. 2008; 283:29273–29284.

8. Wang B, Zhao A, Sun L, Zhong X, Zhong J, Wang H, et al. Protein phosphatase PP4 is overexpressed in human breast and lung tumors. Cell Res. 2008; 18:974–977.

9. Kloeker S, Wadzinski BE. Purification and identification of a novel subunit of protein serine/threonine phosphatase 4. J Biol Chem. 1999; 274:5339–5347.

10. Zhang X, Ozawa Y, Lee H, Wen YD, Tan TH, Wadzinski BE, et al. Histone deacetylase 3 (HDAC3) activity is regulated by interaction with protein serine/threonine phosphatase 4. Genes Dev. 2005; 19:827–839.

11. Lin HC, Wu CL, Chen YL, Huang JS, Wong TY, Yuan K. High-level β1-integrin expression in a subpopulation of highly tumorigenic oral cancer cells. Clin Oral Investig. 2014; 18:1277–1284.

12. Ren W, Wang X, Gao L, Li S, Yan X, Zhang J, et al. MiR-21 modulates chemosensitivity of tongue squamous cell carcinoma cells to cisplatin by targeting PDCD4. Mol Cell Biochem. 2014; 390:253–262.

13. Wang Y, Yuan F. Delivery of viral vectors to tumor cells: extracellular transport, systemic distribution, and strategies for improvement. Ann Biomed Eng. 2006; 34:114–127.

14. Takeshita F, Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006; 97:689–696.

15. Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002; 296:550–553.

16. Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002; 2:243–247.

17. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001; 411:494–498.

18. Rouzier R, Perou CM, Symmans WF, Ibrahim N, Cristofanilli M, Anderson K, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005; 11:5678–5685.

19. Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014; 384:164–172.

20. Toyo-oka K, Mori D, Yano Y, Shiota M, Iwao H, Goto H, et al. Protein phosphatase 4 catalytic subunit regulates Cdk1 activity and microtubule organization via NDEL1 dephosphorylation. J Cell Biol. 2008; 180:1133–1147.

21. Lee DH, Pan Y, Kanner S, Sung P, Borowiec JA, Chowdhury D. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol. 2010; 17:365–372.




