Abstract
Purpose
It is widely accepted that aldehyde dehydrogenase (ALDH) activity is a signature of breast cancer stem cells, and high activity has been reported to be associated with poor clinical outcome. The aim of this study was to assess the expression of members of the ALDH family of isozymes in breast cancer tissues and to evaluate the implications of the results.
Methods
We analyzed paraffin-embedded tumor tissue from 160 patients with breast cancer. Immunohistochemistry (IHC) staining was performed on the slides using antibodies against different ALDH family members. We collated the IHC results with patient clinical characteristics and determined their prognostic value. In addition, we analyzed normal, hyperplastic, and carcinomatous tissues in situ to check their ALDH distributions.
Breast cancers are highly heterogeneous tumors that contain a hierarchy of colonies consisting of cells with stem/progenitor-mature characteristics. The biological properties of the tumor cells in this reservoir are quite different from each other, having distinctly different capacities for invasion, epithelial-mesenchymal transition (EMT), local and distant metastasis, and so forth. It is still unclear which cancer cell groups are highly malignant, with a high probability of invasion, EMT, and metastasis. Cancer stem cells (CSCs) are defined as a subpopulation of cancer cells that have some phenotypic similarities with normal stem cells from adult tissue [1], such as being multipotent for differentiation, having a slow cell cycling time, and occasionally entering a dormant state. They also share a greater probability of undergoing EMT and of acquiring somatic mutations, and are resistant to radiation and chemotherapy, with the resistance level depending on the addition of toxin to the dividing cells [2,3,4]. The identification and quantification of CSCs in breast cancer tissue may therefore be of prognostic significance.
The CD44+/CD24-/low phenotype in breast cancer cells was first identified as a marker for CSCs by Al-Hajj et al. in 2003 [5]. Intensive follow-up studies showed that only a subpopulation of CD44+/CD24-/low cells retain a self-renewal capability [6]. Other markers for breast CSCs were also investigated, including aldehyde dehydrogenase (ALDH) activity, which can be measured using the aldefluor assay. The activity of ALDH is also widely used as a CSCs marker in many types of cancer, e.g., acute myeloid leukemia, as well as solid tumors of the lung, liver, colon, brain, thyroid, bone, pancreas, skin (melanoma), head and neck, prostate, bladder, and cervix [7,8,9,10,11,12,13,14,15,16,17,18,19]. Ginestier et al. [20] reported that aldefluor-positive breast cancer cells were CSCs with self-renewal/differentiation properties and high tumorigenic activity. However, despite the positive association between ALDH 1A1 level and poor clinical outcome, many other breast cancer studies failed to correlate the prevalence of ALDH 1A1-producing cells with tumor grade, metastasis, therapeutic resistance, or clinical outcome [21,22,23]. Marcato et al. [23] reported that, rather than ALDH 1A1, the 1A3, 4A1, 6A1, and 7A1 isoforms were highly expressed in breast cancer, and that 1A3 may be a predictive marker for metastasis.
As there has been no systematic analysis of the utility of the ALDH family in breast cancer prognosis, we searched for highly expressed ALDH family members in breast cancers, monitoring 1A1, 1A3, 4A1, 6A1, and 7A1 expression in 160 patients, and followed their clinical progress and outcomes.
The samples were of human breast neoplasm tissue removed during surgery. Patient anonymity was preserved in all the cases. Approval for the study was granted by the Ethics Committee of West China Hospital (No. 2013-191). We analyzed paraffin-embedded tumor tissue from 160 patients with breast cancer who underwent breast surgery between 2006 and 2009 at West China Hospital. Surgical specimens were obtained before systemic treatment, and paraffin embedding was performed within the framework of diagnostic procedures. Disease-free survival (DFS) and overall survival (OS) were defined as the time between the initial surgery and local or distant metastatic relapse, and between surgery and death, respectively.
From the formalin-fixed paraffin-embedded (FFPE) samples, we prepared tissue microarrays containing cylindrical tissue punches of 1.5 mm diameter. For the prevalence of ALDH 1A1, 1A3, 4A1, 6A1, and 7A1 expression, we used 120 FFPE blocks comprised of 30 normal tissues (sometimes adjacent to neoplasms), 30 hyperplastic tissues, 30 ductal carcinomas in situ (DCIS), and 30 invasive ductal carcinoma (IDC) samples.
The slides were deparaffinized and rehydrated in water. Endogenous peroxidase was blocked with 3% H2O2, and epitope retrieval was performed in a pressure sterilizer. After blocking with 10% serum for 20 minutes at room temperature (RT), the slides were further incubated overnight at 4℃ with the following primary antibodies: rabbit anti-ALDH 1A1 (1:400; Origene, Rockville, USA), mouse anti-ALDH 1A3 (1:400; Origene), rabbit anti-ALDH 4A1 (1:200; Thermo), rabbit anti-ALDH 6A1 (1:300; Origene), and rabbit anti-ALDH 7A1 (1:400; Origene). After 5 phosphate-buffered saline washes, the slides were incubated with horseradish peroxidase-labeled secondary antibody for 30 minutes at room temperature. The slides were then developed using the Dako REAL™ EnVision™ Detection System (DAKO Code K5007; Dako, Glostrup, Denmark).
Hematoxylin and eosin (H&E) and IHC stainings were assessed by light microscopy. The Staff Pathologist at West China Hospital conducted a standard pathological assessment of the tumors from the anonymous patient panel. The status of the patients' estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) was obtained from their pathology reports. HER2 staining was analyzed according to the American Society of Clinical Oncology guidelines. For IHC staining of ALDH 1A1, 1A3, 4A1, 6A1, and 7A1, the percentage of positives among the tumor cells was recorded. To check the IHC results, a semi-quantitative evaluation was carried out in which the percentage (P) of positive cells (score 0 for 0%, 1 for ≤1%, 2 for 1%-10%, 3 for 10%-33%, 4 for 33%-66%, and 5 for 66%-100% positive cells) and the intensity (I) of staining (score 0 for negative, 1 for weak, 2 for moderate, and 3 for strong staining) were included, and a Quickscore was generated. (Q=P+I; score range, 0-8) [24]. For the ALDH 1A1, 1A3, 4A1, 6A1, and 7A1 slides, a Quickscore of 0 to 2 was taken as negative, and a score of 3 or above as positive (Figure 1).
The definitions used for the breast cancer molecular subtypes were as follows: luminal A (ER positive [ER+] and/or PR positive [PR+], and HER2 negative [HER2-]); luminal B (ER+ and/or PR+, HER2+); basal-like (ER-, PR-, HER2-, cytokeratin 5/6 positive and/or HER1+); HER2+/ER- (ER-, PR-, HER2+), and unclassified (negative for all 5 markers).
Statistical analyses were conducted using SPSS version 16.0 software (SPSS Inc., Chicago, USA) with a 5% two-tailed significance level considered statistically significant. Differences in ALDH 1A1, 1A3, 4A1, 6A1, and 7A1 levels in different tissues were analyzed using a rank sum test. Associations between the prevalence of ALDH 1A1-, 1A3-, 4A1-, 6A1-, and 7A1-positive tumor cells and clinical parameters were evaluated with chi-square and Fisher exact tests. Univariate survival analysis was conducted with the Kaplan-Meier method, and multivariate survival analysis was carried out using the Cox proportional hazard model.
All patients were female, ranging in age from 29 to 87 years (mean, 50.7 years). The mean follow-up time was 59.04 months, the mean disease-free survival time was 52.65 months, and the mean overall survival time was 58.8 months. The clinical characteristics studied included histology, grading, tumor size, nodal status, metastasis, clinical stage, ER, PR, HER2/neu, and recurrence. These are listed in Table 1 along with DFS and OS. As expected, nodal metastasis status was found to be significantly correlated with both DFS and OS. Distant metastasis status (when diagnosed) and recurrence were found to be significantly associated with DFS. Also, PR and HER2 statuses were significantly related to both DFS and OS (p<0.05). The better clinical outcomes for PR-positive and HER2-positive patients were considered to be due to the benefits of personalized treatments, such as hormonal therapy for PR-positive patients, and Herceptin® (trastuzumab) treatment for HER2-positive patients.
Expression of ALDH 1A1, 1A3, 4A1, 6A1, and 7A1 was detected with different frequencies in the cytoplasm of epithelial cells in normal, hyperplastic, DCIS, and IDC samples. In Table 2, the average percentage of positive cells for each specific ALDH isoform is listed in the upper row, and the percentage range of positive cells is shown in the lower row. Although the average percentage of positive cells varied between these four groups, we found no significant increase when comparing hyperplastic samples with DCIS, or DCIS with IDC samples. The frequencies of ALDH 1A1, 1A3, 4A1, 6A1, and 7A1 positives in normal, hyperplastic, DCIS, and IDC tissues are detailed in Table 2.
To learn of any correlations between the levels of the ALDH enzyme family and clinical characteristics, we analyzed the expression of all the family members in the clinical specimens. Because of deterioration in the quality of some slides following IHC, we were only able to analyze 130, 129, 132, 133, and 141 samples for ALDH 1A1, 1A3, 4A1, 6A1, and 7A1, respectively. We found no significant correlation between ALDH 1A1, 4A1, 6A1, and 7A1 and clinical characteristics, e.g., histology, grading, tumor size, nodule status, distant metastasis (initial metastasis was excluded), clinical stage, ER status, PR status, HER2 status, as well as local or distant recurrences (data not shown). For ALDH 1A3, however, we found that the level of expression was significantly correlated with distant metastasis of breast cancer (p=0.001), but there were no significant correlations with the other clinical characteristics (Table 3). Of the 37 breast cancer patients who subsequently developed distant metastases, 32 of the tumors (86.49%) were found to have a high prevalence of ALDH 1A3-expressing cells in the initial tumor resection.
To further evaluate the prognostic value of the expression of ALDH family members in tumor cells, we looked for correlations between the levels of the different ALDH isoforms and DFS and OS. As shown in Table 4, only ALDH 1A3 expression was found to correlate with both DFS (p<0.001) and OS (p=0.002). Expression of the other isoforms was not significantly correlated with DFS or OS (data not shown), and no significant association was observed between the prevalence of ALDH 1A3 and that of the other molecular subtypes in our study (Table 5). Data from the analysis of the prognostic potential of ALDH 1A3 expression are detailed in Figure 2. We observed a clear and significant difference in DFS and OS between ALDH 1A3-positive and ALDH 1A3-negative groups of patients. Based on our 87-month follow-up study, expression of ALDH 1A3 correlates with distant metastasis, DFS, and OS. The expression of ALDH 1A3 in breast tumor cells can thus be considered a predictor of poor clinical outcomes in breast cancer.
Breast cancer is currently the most common cancer among women in the United States [25]. A better understanding of the biological properties of cancer cells is likely to benefit breast cancer patients in areas such as cancer risk assessment, treatment, and prognostication. There are many tests available to predict clinical outcomes when breast cancer is first diagnosed, e.g., the Oncotype DX® and MammaPrint tests. These assays are based on measuring the expression of tens of genes with no histological considerations. Because breast cancer cells are quite heterogeneous neoplasia, the biological properties as well as the genetic background of cells in the same tumor might be quite different from each other. In this study, we tried to identify a subgroup of cells within the breast tumor with high invasive ability, resistance to treatment, EMT, and metastatic potential. We assume that this subgroup of cells significantly contributes to breast cancer recurrence, metastasis, and poor clinical outcome.
The recently identified CSCs are a subpopulation of cancer cells that have some phenotypic similarities with stem cells in adult tissues [1,2,3,4]. Although the concept of breast CSC may be challenged, there is a subgroup of cells within breast tumors that are multipotent for differentiation, exhibit slow cell cycling and even, occasionally, a dormant status, are resistant to radiation and chemotherapies, and have a high potential for EMT [1]. ALDH activity, which can be measured using the aldefluor assay, is widely considered as a CSC marker, not only for breast cancer, but also for cancers from other sources [7,8,9,10,11,12,13,14,15,16,17,18,19]. Translating this concept into clinical practice, however, requires practical techniques such as immunohistochemistry. The current problem is knowing which member(s) of the ALDH gene family of isoforms contribute to the measured ALDH activity and are suitable for prognostic purposes.
The ALDH gene family has 19 members, and their distribution varies depending on the tissue in which they occur. Although ALDH 1A1 has been reported as being highly expressed in many tissues and contributing to ALDH activity, this may not be true in breast tissue [26]. It was previously reported that the 1A3, 4A1, 6A1, and 7A1 isoforms of ALDH, and not the 1A1 form, were highly expressed in breast cancer. As there had been no systematic analysis of ALDH isoforms and their relationship to the clinical character and outcome of breast cancer, we measured their expression and searched for correlations with clinical characteristics and disease outcomes. Consistent with literature stating that only ALDH 1A3 contributes to aldefluor activity in breast cancer, we found that only the 1A3, 4A1, 6A1, and 7A1 isoforms were highly expressed in breast cancer tissues, and of these, only the 1A3 level was found to be significantly correlated with distant metastasis (p=0.001), DFS (p<0.001), and OS (p=0.001).
As it had previously been shown that ALDH 1A3 is the primary contributor to aldefluor activity in breast cancer tissue [23], we further investigated the relationship of ALDH 1A3 with breast cancer gene expression subtypes, as these have been widely reported to correlate both with prognosis and choice of treatment regimen. We found no significant association of ALDH 1A3 expression with certain molecular subtypes of breast cancer. We speculate that measuring a single characteristic of tumors might not be sufficient to predict the likelihood of their progression, distant metastasis, and recurrence, or the eventual clinical outcome. This is because tumor progression is a complex biological process, involving the contributions of multiple types of cells as well as microenvironmental issues. Our data are partially consistent with previous data in suggesting that the expression of ALDH 1A3 is likely an immuno-phenotype that is independent of the status of ER, PR, and HER2.
Our systematic analysis of highly expressed ALDH family members in breast cancer tissue showed that ALDH 1A3 is a predictive marker of poor clinical outcomes. Further studies could focus on the role of CSCs in distant metastasis and progress in EMT.
Figures and Tables
Figure 1
Typical images of immunihistochemistry (IHC) staining for aldehyde dehydrogenase (ALDH) 1A3 expression in invasive breast carcinoma (IHC stain for ALDH 1A3, ×400). (A) Absence of ALDH 1A3 expression in tumor (score=0). (B) Few ALDH 1A3 tumor cells with moderate staining intensity (score=1+2; P+I). (C) Abundant of ALDH 1A3 tumor cells with moderate staining intensity (score=2+4; P+I). (D) Abundant of ALDH 1A3 tumor cells with strong staining intensity (score=3+5; P+I).
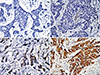
Figure 2
The prevalence of aldehyde dehydrogenase (ALDH) 1A3 positive tumors cells with the clinical outcomes. Univariate survival analysis of ALDH 1A3 expression was performed in disease-free survival (A) and overall survival (B).

Table 3
Prevalence of ALDH 1A3 tumor cells in breast tumors stratified according to clinical characteristics

Notes
References
2. Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010; 107:18115–18120.

3. Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea: a paradigm shift. Cancer Res. 2006; 66:1883–1890.
4. Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011; 121:3804–3809.

5. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003; 100:3983–3988.

6. Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008; 10:R25.

7. Basak SK, Veena MS, Oh S, Huang G, Srivatsan E, Huang M, et al. The malignant pleural effusion as a model to investigate intratumoral heterogeneity in lung cancer. PLoS One. 2009; 4:e5884.

8. Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008; 6:1146–1153.

9. Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, et al. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009; 69:8208–8215.

10. Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009; 69:3382–3389.

11. Rasper M, Schäfer A, Piontek G, Teufel J, Brockhoff G, Ringel F, et al. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro Oncol. 2010; 12:1024–1033.

12. Todaro M, Iovino F, Eterno V, Cammareri P, Gambara G, Espina V, et al. Tumorigenic and metastatic activity of human thyroid cancer stem cells. Cancer Res. 2010; 70:8874–8885.

13. Wang L, Park P, Zhang H, La Marca F, Lin CY. Prospective identification of tumorigenic osteosarcoma cancer stem cells in OS99-1 cells based on high aldehyde dehydrogenase activity. Int J Cancer. 2011; 128:294–303.

14. Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010; 102:340–351.

15. Boonyaratanakornkit JB, Yue L, Strachan LR, Scalapino KJ, LeBoit PE, Lu Y, et al. Selection of tumorigenic melanoma cells using ALDH. J Invest Dermatol. 2010; 130:2799–2808.

16. Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010; 32:1195–1201.

17. Li T, Su Y, Mei Y, Leng Q, Leng B, Liu Z, et al. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients' outcome. Lab Invest. 2010; 90:234–244.

18. Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z, et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010; 19:327–337.

19. Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010; 5:e10277.

20. Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007; 1:555–567.

21. Neumeister V, Rimm D. Is ALDH1 a good method for definition of breast cancer stem cells? Breast Cancer Res Treat. 2010; 123:109–111.

22. Resetkova E, Reis-Filho JS, Jain RK, Mehta R, Thorat MA, Nakshatri H, et al. Prognostic impact of ALDH1 in breast cancer: a story of stem cells and tumor microenvironment. Breast Cancer Res Treat. 2010; 123:97–108.

23. Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011; 29:32–45.

24. Currie MJ, Beardsley BE, Harris GC, Gunningham SP, Dachs GU, Dijkstra B, et al. Immunohistochemical analysis of cancer stem cell markers in invasive breast carcinoma and associated ductal carcinoma in situ: relationships with markers of tumor hypoxia and microvascularity. Hum Pathol. 2013; 44:402–411.

25. American Cancer Society. Cancer Treatment & Survivorship Facts & Figures 2012-2013. Atlanta: American Cancer Society;2012.
26. Rexer BN, Zheng WL, Ong DE. Retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in MCF-7 cells. Cancer Res. 2001; 61:7065–7070.




 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download