Abstract
Purpose
Genetic variation in fibroblast growth factor receptor 2 (FGFR2) is a newly described risk factor for breast cancer. This study aimed to evaluate the association of four single nucleotide polymorphisms (SNPs) in FGFR2 with breast cancer in Han Chinese women.
Methods
Two hundred three women with breast cancer and 200 breast cancer-free age-matched controls were selected. Four SNPs (rs2981579, rs1219648, rs2420946, and rs2981582) and their haplotypes were analyzed to test for their association with breast cancer susceptibility. The presence of the four FGFR2 SNPs was determined by polymerase chain reaction-restriction fragment length polymorphism analysis.
Results
A statistically significant difference was observed in the frequency of rs2981582 in the FGFR2 gene (p<0.05) between case and control groups. In subjects stratified by menopausal status, rs2981582 TT, rs2420946 AA, and rs1219648 CC were significantly associated with the risk of breast cancer in postmenopausal subjects, but no significant associations between these four SNPs and the risk of breast cancer were identified in premenopausal subjects. Further, there was no significant association between hormone receptor status (estrogen receptor and progesterone receptor) and breast cancer risk. Six common (> 3%) haplotypes were identified. Three of these haplotypes, CGTC (odds ratio [OR], 0.613; 95% confidence interval [CI], 0.457-0.82; p=0.001), TGTC (OR, 6.561; 95% CI, 2.064-20.854; p<0.001), and CATC (OR, 12.645; 95% CI, 1.742-91.799; p=0.001) were significantly associated with breast cancer risk.
Breast cancer is the most commonly occurring cancer and the leading cause of cancer death among women worldwide, accounting for 23% of all cancers and 400,000 deaths annually [1]. Although numerous genetic association studies on Breast cancer have been published, few candidate genes for this disease have been identified, such as HER2, CyclinD1, BRCA1/2, etc. Two genome-wide association (GWA) studies reported that five single nucleotide polymorphisms (SNPs) (rs11200014, rs2981579, rs1219648, rs2420946, and rs2981582) within fibroblast growth factor receptor 2 (FGFR2) were associated with breast cancer risk [2,3]. However, this association was restricted to SNPs in the linkage disequilibrium (LD) block of intron 2. Both studies had similar findings, demonstrating that all five SNPs were in high LD. However, efforts to identify the causal allele by resequencing the FGFR2 intron 2 region and fine-mapping this region with haplotype-tagging SNPs have not yet revealed the causal sequence variant in European and Asian populations [4].
FGFR2 is a member of the receptor tyrosine kinase family, which includes distinct fibroblast growth factor receptors involved in tumorigenesis. The association of FGFR2 polymorphisms with breast cancer has also been reported in Jewish and Arab Israeli [5], Taiwanese [6], European-American, and African-American populations [7,8]. The mechanism underlying how these intronic SNPs increase the risk of breast cancer remains unknown, although an association between these SNPs and reproductive hormones has been suggested in an epidemiological study that described a significant association between FGFR2 intronic polymorphisms and the efficacy of combined hormone replacement therapy in European-American women [7].
FGFR2 is often overexpressed in breast cancer [9-11], and data suggest that FGFR2 acts as a tumor suppressor in prostate and urothelial cancers [12]. The aberrant expression of alternatively spliced isoforms of FGFR2 transforms breast cancer cells through sustained signal transduction, thoes results suggest that different splice variants have differing transforming activities [9-13]. The present case-control study investigated these loci using a large number of samples of the same ethnic origin to verify the putative association between FGFR2 polymorphisms and breast cancer.
Further to the important role of FGFR2 in breast carcinogenesis and the previously published association of FGFR2 polymorphisms with various malignant diseases, we sought to estimate the contribution of FGFR2 polymorphisms to breast cancer risk in Han Chinese women.
Breast cancer was histopathologically confirmed in 203 women with an average age was 48.6±10.1 years at the General Hospital of Ningxia Medical University. This study was approved by the Ethics Committee of the hospital (No. 2013-172, Ningxia Medical University Medical Ethical Committee, China), and all participants provided their written informed consent for blood sampling and genetic testing. Clinicopathological data, including estrogen receptor (ER) and progesterone receptor (PR) status, were collected, and patients with a previous history of cancer and treatment with chemotherapy or radiotherapy were excluded from the study.
To determine the menopausal status of study subjects at each visit, we used the menopausal status algorithm developed by the Women's Ischemia Syndrome Evaluation study [14]. This algorithm uses self-reported reproductive history and menstrual cycle pattern and dates in combination with serum reproductive hormone levels to evaluate menopausal status in cases where a one-time serum sample is drawn without consideration of the specific day of the menstrual cycle. Importantly, in addition to using this algorithm, for women who were not clearly postmenopausal, we also analyzed all reproductive and relevant biometric information and classified both cases and controls according to their menopausal status.
The 200 cancer-free controls were randomly selected from a pool of healthy volunteers who visited the health check-up center at the General Hospital during the same period, and were age-matched with the cancer cases (±5 years). Female controls who responded to questionnaire, without a history of breast cancer and with no indication of breast cancer on a baseline mammogram and a clinical breast examination at the time of blood collection, were included in the study. Demographic and breast cancer-related information was obtained by personal interviews. All cases and controls were from the Ningxia region and had lived there for more than 10 years. The clinical information collected included age at menarche, age at menopause, age at the first live birth and the total months of breastfeeding among parous women, oral contraceptive use, hormone replacement therapy use, family history of breast cancer, and prior diagnosis of benign breast disease. For all patients and controls who had fasted overnight, two 5-mL blood samples were collected in ethylenediaminetetraacetic acid-coated tubes and stored at -80°C until DNA was extracted. All samples were coded in order to insure their anonymity.
Genomic DNA was extracted from peripheral blood samples using a Genomic DNA extraction Kit (SBS Genetech Ltd., Corp., Beijing, China). FGFR2 was amplified by polymerase chain reaction (PCR); the primer sequences, designed using Primer Premier 5 software (Songon Biotechnology Ltd., Corp., Shanghai, China), are described in detail in Table 1. PCR was performed in a 50-µL reaction mixture containing 50 ng DNA, 0.1 µM of each primer, 0.5 units Taq DNA polymerase, and 25 µL 2X SuperMix. Standard PCR cycling conditions were employed: predenaturation at 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C to 65°C for 30 seconds, and extension at 72°C for 30 seconds, with a final extra extension at 72°C for 10 minutes. PCR amplicons were digested with restriction enzymes (Table 1). The digestion products were then electrophoresed on a 3% agarose gel and observed using the Gel Doc 2000 system (Bio-Rad, Hercules, USA). Pictures of agarose gels for PCR and restriction fragment length polymorphism are shown in Figure 1.
Where appropriate, chi-square analysis was performed. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated accordingly. A p-value of <0.05 was considered statistically significant. For statistical analysis, the SPSS statistical software system version 13.0 (SPSS Inc., Chicago, USA) was used.
The Hardy-Weinberg equilibrium was tested for the FGFR2 SNP genotypes in patients with breast cancer and in controls. SHEsis online software (http://analysis.bio-x.cn/myAnalysis.php, The Bio-X Research Institute of Shanghai Jiao Tong University, Shanghai, China) was used to construct the haplotypes for the identified polymorphisms. Unadjusted ORs (95% CI) and the respective p-values are shown in Table 2. Significant associations were found between rs2981582 TT and the risk of breast cancer, but no significant associations were ascertained between the risk of breast cancer and the other three SNPs in FGFR2.
Table 2 shows the genotype distributions of the four SNPs in subjects stratified by menopausal status. In postmenopausal subjects, rs2981582 TT, rs2420946 AA, and rs1219648 CC were significantly associated with the risk of breast cancer, but no significant associations were found between the four SNPs and risk of breast cancer in premenopausal subjects.
Clinicopathological characteristics (ER and PR status) were not associated with breast cancer risk (Table 3). Haplotype analysis identified six common haplotypes with frequencies greater than 0.03 (Table 4). Of these haplotypes, three, CGCT (OR, 0.61; 95% CI, 0.46-0.81; p=0.001), TGTC (OR, 6.56; 95% CI, 2.06-20.85; p<0.001), and CATC (OR, 12.65; 95% CI, 1.74-91.80; p=0.001), were significantly associated with breast cancer risk.
The present study estimates the contribution of FGFR2 polymorphisms to breast cancer risk in Han Chinese women. For this purpose, we genotyped four SNPs (rs2981579, rs1219648, rs2420946, and rs2981582) in intron 2 of FGFR2 that were found to be most significantly associated with breast cancer in a previous GWA study. The study by Easton et al. [2] showed that four SNPs in intron 2 of the FGFR2 gene (rs11200014, rs2981579, rs1219648, and rs2420946) are strongly associated with postmenopausal breast cancer in 1,145 cases and 1,142 age-matched controls. The most frequent risk haplotype (AAGT) was found to be significantly associated with breast cancer risk in all populations. Another large three-stage GWA study of 4,398 breast cancer cases and 4,316 controls identified six SNPs in five novel independent loci that were associated with breast cancer, and this finding was confirmed in an additional 21,860 cases and 22,578 controls in 22 studies [5]. The most significantly associated SNP, rs2981582, lies within a 25-kb LD block in intron 2 of FGFR2 and had an r2 of 1.0 with rs1219648 and rs2420946, 0.97 with rs2981579, and 0.96 with rs11200014 based on the HapMap Central European-like Utahns samples. The high degree of LD indicates a close correspondence between the results of both studies, both of which genotyped white non-Hispanic women of European descent. Four of 22 validation studies in the larger GWA study included women of Asian descent. The association of SNPs in intron 2 of FGFR2 with breast cancer risk was essentially confirmed in all populations participating in these GWA studies, but with different degrees of risk for European and Asian populations. The study by Raskin et al. [5] confirms this strong and consistent association, extends these results to Ashkenazi and Sephardi Jews, and appears to suggest a modest risk in Arab populations as well. Genetic variation in FGFR2 appears to be implicated in a considerable proportion of breast cancer cases in Israel and throughout the world.
Our results suggest that the rs2981582 TT genotype is significantly associated with breast cancer in Chinese women (p<0.05), but no associations were detected between the risk of breast cancer and the other three SNPs (rs2420946, rs1219648, and rs2981579) in FGFR2. In subjects stratified by menopausal status, rs2981582 TT, rs2420946 AA, and rs1219648 CC genotypes were significantly associated with the risk of breast cancer in postmenopausal subjects, but no evidence of significant associations were found in premenopausal subjects. Although the LD pattern observed in our sample is similar to that observed in HapMap and other studies, the magnitude of LD in intron 2 of FGFR2 generally seems to be lower in our population when compared with that in the HapMap.
A study by Liang et al. [15] showed that the associations between the risk of breast cancer and SNP rs2981582C/T, rs1219648A/G, and rs2420946C/T appeared to be strongest in premenopausal women and those with ER/PR-positive tumors, indicating that these variant genotypes may be involved in the etiology of breast cancer through pathways related to estrogen and/or progesterone. However, clinicopathological characteristics (ER and PR status) were not significantly associated with breast cancer risk in the Han Ningxia population in this study. Many study indicated that genes expressed differentially between the samples from different ethnic population, therefore, there may be a variety of racial and ethnic populations in different studies, depending on the population structure. Besides, some limitations should be considered when interpreting the results of this study such as, small sample sizes largely impact on the accuracy and precision of results, so the importance of studying very large sample sizes to achieve sufficient statistical power in order to confidently evaluate the possible effect interactions should be considered in future studies.
By haplotype analysis, we identified six common haplotypes with frequencies greater than 0.03. Three common risk haplotypes, CGCT, TGTC, and CATC, were significantly associated with breast cancer risk. The causal allele associated with these risk haplotypes may reveal the functional variant underlying changes in FGFR2 function. Meyer et al. [16] have recently shown that the haplotype marked by the minor allele of rs2981582 is associated with a higher level of FGFR2 transcription both in cell lines and in tumors. The authors hypothesized that this could lead to an alteration in the transcription level of the corresponding allele. Further functional studies are required to elucidate the actual causative variant located within intron 2 of the FGFR2 gene.
In summary, we have shown that rs2981582 and the haplotypes of four SNPs within the LD block in intron 2 of FGFR2 are associated with breast cancer risk in Han Chinese women.
Figures and Tables
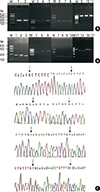 | Figure 1Results of polymerase chain reaction (PCR)-restriction fragment length Polymorphism (RFLP) and sequence analysis carried out for resolution of FGFR2 rs2981582, rs2420946, rs1219648 and rs2981579. (A) The amplification of PCR products for four FGFR2 SNPs. Lanes 1-2 (176 bp), lanes 3-5 (429 bp), lanes 6-8 (230 bp) and lanes 9-11 (437 bp), Lane M: 100-bp ladder marker. (B) The genotyping for rs2981582, rs2420946, rs1219648, and rs2981579 of FGFR2. Lanes 1-3 of rs2981582 genotype patterns, TT[176 base pairs (bp)], TC (176+154+22bp), CC (154+22 bp); Lanes 4-6 of rs2420946 genotype patterns, GG (321+108bp), AG (429+321+108 bp), AA (429bp); Lanes 7-10 of rs1219648 genotype patterns,TT (230 bp) (7-8), CC (211+19 bp), TC (230+211+19 bp); Lanes 11-17 of rs rs2981579 genotype patterns, TT (437 bp) (lanes 11, 15, 16), CC (350+87 bp) (lanes 12, 13, 14), TC (437+350+87 bp) (lane 17), and Lane M:100-bp ladder marker. (C) Results of DNA sequence analysis, Wild and Mutant allele of four SNPs are indicated with arrows, respectively. |
Table 2
The genotypic frequencies of FGFR2 SNPs between breast cancer and female controls and stratified analysis by menopausal status

FGFR2=fibroblast growth factor receptor 2; SNP=single nucleotide polymorphism; OR=odds ratio; CI=confidence interval.
*Significant association between risk of breast cancers and controls (p<0.05). Adjusted for reference age and menopausal status. There was no difference between the crude ORs and adjusted ORs.
Table 3
Stratified analyses between combined effects of FGFR2 and breast cancer risk by age, condition of lymphatic metastasis, and ER/PR status

Notes
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61:69–90.

2. Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007; 447:1087–1093.
3. Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007; 39:870–874.

4. Heath SC, Gut IG, Brennan P, McKay JD, Bencko V, Fabianova E, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008; 16:1413–1429.

5. Raskin L, Pinchev M, Arad C, Lejbkowicz F, Tamir A, Rennert HS, et al. FGFR2 is a breast cancer susceptibility gene in Jewish and Arab Israeli populations. Cancer Epidemiol Biomarkers Prev. 2008; 17:1060–1065.

6. Lin CY, Ho CM, Bau DT, Yang SF, Liu SH, Lin PH, et al. Evaluation of breast cancer susceptibility loci on 2q35, 3p24, 17q23 and FGFR2 genes in Taiwanese women with breast cancer. Anticancer Res. 2012; 32:475–482.
7. Rebbeck TR, DeMichele A, Tran TV, Panossian S, Bunin GR, Troxel AB, et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009; 30:269–274.

8. Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010; 31:1417–1423.

9. Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005; 16:179–186.

10. Moffa AB, Ethier SP. Differential signal transduction of alternatively spliced FGFR2 variants expressed in human mammary epithelial cells. J Cell Physiol. 2007; 210:720–731.

11. Moffa AB, Tannheimer SL, Ethier SP. Transforming potential of alternatively spliced variants of fibroblast growth factor receptor 2 in human mammary epithelial cells. Mol Cancer Res. 2004; 2:643–652.
12. Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005; 16:139–149.

13. Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001; 61:3541–3543.
14. Johnson BD, Merz CN, Braunstein GD, Berga SL, Bittner V, Hodgson TK, et al. Determination of menopausal status in women: the NHLBI-sponsored Women’s Ischemia Syndrome Evaluation (WISE) Study. J Womens Health (Larchmt). 2004; 13:872–887.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download