Abstract
Purpose
The DNA methylation mediated by specific DNA methyltransferases (DNMTs), results in the epigenetic silencing of multiple genes which are implicated in human breast cancer. We hypothesized that the natural compounds modulate the expression of DNMTs and their associated proteins in the breast cancer cell lines and affect the methylation mediated gene silencing.
Methods
The DNMTs transcript expression was analyzed by reverse transcription-polymerase chain reaction (RT-PCR) in the tumors and the adjacent normal breast tissues of the patients with invasive ductal breast carcinoma. We tested the hypothesis that the natural compounds, viz., epigallocatechin gallate (EGCG), genistein, withaferin A, curcumin, resveratrol, and guggulsterone, have demethylation potential. To investigate this hypothesis, we analyzed the DNMTs expression at the transcript levels, followed by the analysis of DNMT1 and its associated proteins (HDAC1, MeCP2, and MBD2).
Results
The increased DNMTs transcripts expression, viz., DNMT1, DNMT3a, and DNMT3b, in the breast cancer tissues suggest involvement of the DNMTs in the breast carcinogenesis. Quantitative RT-PCR analysis revealed that the treatment with natural compounds, viz., EGCG, genistein, withaferin A, curcumin, resveratrol, and guggulsterone, resulted in a significant decrease in the transcript levels of all the DNMTs investigated. Importantly, these natural compounds decreased the protein levels of DNMT1, HDAC1, and MeCP2.
Conclusion
Our results demonstrate that the natural compounds, EGCG, genistein, withaferin A, curcumin, resveratrol, and guggulsterone, have the potential to reverse the epigenetic changes. Moreover, their lack of toxicity makes these natural compounds promising candidates for the chemoprevention of the breast cancer. In-depth future mechanistic studies aimed to elucidate how these compounds affect the gene transcription are warranted.
DNA hypermethylation plays an important role in silencing the tumor suppressor genes being one of the most consistent hallmarks of the human cancers, and the phenomenon is of a comparable significance to classic genetic mutations [1]. In recent years, DNA methylation has emerged as an attractive target for the cancer therapeutics [2]. DNA methylation is catalyzed by a family of enzymes called DNA methyltransferases (DNMTs) [3]. The human genome contains four DNA methyltransferase genes, DNMT1, DNMT2, DNMT3A, and DNMT3B which encode proteins with distinct functional specificities. Among these DNMTs, DNMT1 is the most abundant DNA methyltransferase in the mammalian cells, and considered to be the key maintenance methyltransferase in the mammals. It has been established that the inhibition of DNA methyltransferase activity can strongly inhibit the formation of tumors [4]. The repressive effects of DNA methylation are mediated in large part by the methyl-CpG binding proteins (MCBPs) and also associated with histone modifications. MCBPs, such as MeCP2, methyl-CpG binding domain 1 (MBD1), and MBD2, specifically bind to CpG methylated DNA and are associated with the histone deacetylase (HDAC)-containing complexes, to "erase" the transcription-activating histone acetyl marks.
Natural products have received increasing attention in the recent years as novel anticancer agents [5-9]. Interest in the potential cancer chemopreventive and therapeutic properties of the diet-derived compounds, including those of the plant polyphenols, has increased tremendously. These compounds can be found in many fruits and vegetables including soya, turmeric, grapes, celery, apples, onions, parsley, capsicum, green tea, pepper, etc. and have been shown to possess anticancer activities [10]. The mechanisms by which the flavonoids exert the anticancer effects are varied and may include action through anti-inflammation [11], free radical scavenging [12], modulation of survival and proliferation pathways [13,14], and inhibition of the ubiquitin-proteasome pathway [15,16]. The potential of the nutraceutical agents in combination therapies is being increasingly considered based on the the findings of the improved animal model outcome when these compounds are combined with radiotherapy and chemotherapy [17].
The primary rationale of this work was to explore the effects of the natural compounds, viz., epigallocatechin gallate (EGCG), genistein, withaferin A, curcumin, resveratrol, and guggulsterone, on the hypermethylation of specific genes and determine their inhibitory effects on the key proteins involved in the DNA hypermethylation mechanism.
Surgically resected tissue samples were collected from the untreated primary breast carcinoma patients (n=40), along with the paired normal breast tissues (n=10) (taken 5-10 cm away from the site of the tumor). The patients were enrolled as outpatients at the Department of Surgical Disciplines, All India Institute of Medical Sciences, New Delhi, India between 2004 and 2008, following the study approval by the Institutional Human Ethics Committee. Written consent was obtained from all patients enrolled in the study. The patient age ranged 30 to 81 years in age (median, 50 years). All patients were diagnosed with invasive ductal carcinoma (IDC) of the breast. A section of each tumor and a matched normal breast tissue were sampled and stored in formalin for the histopathological characterization in the diagnosis conformation and immunohistochemistry. The rest of the tissues were immediately snap frozen and stored at -80℃ for further use. The clinicopathological characteristics of the patients analyzed in this study are summarized in Table 1.
EGCG, genistein, curcumin, resveratrol, guggulsterone, 5-aza-2´-deoxycytidine (Decitabine) and MTT 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide were purchased from Sigma Chemical Co. (Bangalore, India). Curcumin, resveratrol, and guggulsterone were dissolved in dimethyl sulfoxide (DMSO) and stored in dark at -20℃ while genistein, EGCG and withaferin A were dissolved in water and stored at 4℃. Decitabine was dissolved in PBS buffer and stored as a 1 mM solution at -20℃.
Human breast carcinoma cell lines MCF7 and MDA MB 231 were obtained from the American Type Culture Collection (Manassas, USA) and cultured in Dulbecco's Modified Eagle Medium (DMEM). The effect of the natural compounds on cellular proliferation was assessed by MTT assay, according to standard protocols. Briefly, 104 cells were seeded per well in a 96-well plate. After 24 hours of preincubation in their respective media containing 2% FBS, the tested natural compounds were added to the culture medium and the cells were incubated for 96 hours. Control cells were treated with DMSO only, where the concentration of DMSO did not exceed 0.2%. Media were replenished after every 48 hours in both the treated and control cells. After 96 hours, the cells were washed twice with PBS, and a fresh medium containing MTT (0.5 mg/mL) was added. After 4-hour incubation, the formazan crystals were dissolved in acidic isopropanol and the absorbance was measured at 540 nm. All experiments were repeated three times, with at least three measurements (triplicates).
Isolation of genomic DNA and treatment with sodium bisulfite were done according to protocols optimized in our laboratory. The MSP of genes: ERα, PRB, RARβ2, TMS1, Cyclin D2, MGMT, SLIT2, and Maspin were carried out using the primers and conditions as described previously. The details are given in Table 2 [18-20]. All PCRs were performed with positive controls for both the unmethylated and methylated alleles, and with no DNA to check for contamination.
Real-time reverse transcription-polymerase chain reaction (RT-PCR) for DNMT1, DNMT3a and DNMT3b, was done with cDNA synthesized from the total RNAs of the control and the drug-treated cells as described using Syber Green Quantitative-RTPCR kit (Stratagene, Vancouver, Canada) following the manufacturer's instructions. The primer sequences are provided in Table 3. The comparative Ct method was used to calculate the relative changes in gene expression using 7500 Fast Real Time PCR System (Applied Biosystems, Foster City, USA). The relative changes of gene expression were calculated using the following formula: Fold change in gene expression, 2-ΔΔCt=2-[ΔCt (natural compound treated samples)-ΔCt (untreated control)], where ΔCt=Ct (detected genes)-Ct (18S rRNA) and Ct represent the threshold cycle number.
Cells were lysed on ice using lysis buffer (0.05 mol/L Tris-HCl, pH 7.4, 0.15 mol/L NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mmol/L ethylenediaminetetraacetic acid, 0.5 mmol/L dithiothreitol, 1 mmol/L phenylmethylsulfonyl fluoride, 5 mg/mL leupeptin, and 10 mg/mL aprotinin). The lysates were then centrifuged at 13,000 rpm at 4℃ for 10 minutes. Protein extracts were solubilized in sodium dodecyl sulfate (SDS) gel loading buffer (60 mmol/L Tris base, 2% SDS, 10% glycerol, and 5% β-mercaptoethanol). Samples containing equal amounts of protein (80 µg) were separated on an 8% SDS-polyacrylamide gel electrophoresis and electroblotted onto Immobilon-P membranes (Millipore, Bedford, USA) in a transfer buffer. Immunoblotting was performed using antibodies against p21 (1:500; Santa Cruz Biotechnology, Santa Cruz, USA), DNMT1 (1:1,000), HDAC1 (1:2,000), MeCP2 (1:3,000), MBD2 (1:2,000) (Santa Cruz Biotechnology, Billerica, USA), and anti-β-actin antibodies (1:5,000; Santa Cruz), as internal control. The signal was developed with enhanced chemiluminescence (Pierce, Rockland, USA) after incubation with appropriate secondary antibodies.
Statistical analysis was performed using the SPSS version 16.0 statistical software (SPSS Inc., Chicago, USA). The relationships between the expression of DNMTs and the clinicopathological variables were tested using chi-square test. The strength of the association between the expression levels of each DNMT in different sample categories was calculated by the Spearman rank-correlation coefficient.
All breast carcinoma patients included in this study showed amplicons of DNMT1, DNMT3a, and DNMT3b, respectively, as shown in Figure 1. The level of expression of DNMTs in the breast cancer patients was compared, after normalization with β-actin, with the adjacent normal breast tissues. The levels of DNMT1, DNMT3a, and DNMT3b mRNA were observed to be 1.2- to 4.4-folds, 1.1- to 3.77-folds, and 1.06- to 4.01-folds elevated in breast cancer tissues, respectively, as compared to the adjacent normal breast tissues.
In summary, DNMT1 and DNMT3b showed moderately higher transcript levels (mean±SE, 2.9±0.2 and 2.6±0.1, respectively) as compared to DNMT3a (mean±SE, 2.0±0.1). To elucidate the effects of the DNMTs expression on the hormonal receptor status, the correlation was determined between the hormone receptor status and the DNMT expression level. A moderate level of correlation was observed between ER/PR and DNMT1 (r=0.5, p=0.01), DNMT3a (r=0.4, p=0.05), and DNMT3b (r=0.5, p=0.001). Further, the transcript levels were analyzed to determine whether these three DNMTs were expressed coordinately or independently. The mRNA levels of these three DNMTs were observed to be moderately correlated with each other as follows: DNMT1 with DNMT3a (r=+0.6, p=0.001), DNMT1 with DNMT3b (r=+0.6, p≤0.001), and DNMT3a with DNMT3b (r=+0.5, p=0.004).
The effects of the various natural compounds on the cell viability after 96 hours of exposure were assessed by MTT assay. The IC50s for MCF 7 cells were 10, 15, 10, 10, 8, and 20 µM, respectively for EGCG, genistein, curcumin, resveratrol, withaferin A, and guggulsterone (Figure 2). MDA MB 231 cells were more sensitive to genistein and guggulsterone than MCF 7. The IC50s for MDA MB 231 cells were 15, 10, 10, 15, 10, and 15 µM, respectively for EGCG, genistein, curcumin, resveratrol, withaferin A, and guggulsterone (Figure 2).
For evaluation of the ability of the tested natural compounds to reactivate the silenced genes, the methylation status of the promoter regions of ERα, PRB, TMS1, Cyclin D2, MGMT, and Maspin genes in MDA MB 231 and RARβ2 and SLIT2 in MCF 7 cells were assessed (Figure 3). The panel of genes studied was selected on the basis of the involvement in the breast tumor initiation, progression, and metastasis [21,22]. Decitabine was used as the reference compound in all the experiments. MDA MB 231 and MCF 7 cells were treated with decitabine at 6, 8, 10, and 12 µM concentrations for 72 hours and 96 hours to assess the effects on the methylation status in our panel of genes (data not shown). MSP of the panel of genes, ERα, PRB, RARβ2, TMS1, Cyclin D2, MGMT, SLIT2, and Maspin were carried out after treatment of MCF 7 and MDA MB 231 cells with decitabine at the above-mentioned dose and time. Complete demethylation of our panel of genes was observed at 10 µM (96 hours) in MDA MB 231 cells and at 12 µM (96 hours) in MCF 7 cells, while partial demethylation was observed at the above doses at 72 hours. Hence, MDA MB 231 cells treated with 10 µM decitabine and MCF7 cells treated with 12 µM decitabine for 96 hours were used as positive controls for further experiments.
EGCG and genistein, known nonnucleoside demethylating agents showed demethylation of the selective genes only. EGCG treated MDA MB 231 cells showed complete demethylation of RARβ2 and TMS1, whereas EGCG treated MCF 7 cells showed complete demethylation of RARβ2. Genistein treated MDA MB 231 cells showed complete demethylation of Cyclin D2. However, partial demethylation was observed in Cyclin D2 and MGMT on the treatments with EGCG, whereas MGMT was partially demethylated on treatment with genistein. However, curcumin, resveratrol, withaferin A, and guggulsterone had no effect on the methylation status of these genes.
The effect of the tested natural compounds on the DNMTs expression was determined by the quantitative analysis of mRNA for each of the three DNMTs. After treatment with various polyphenols, there was a marked decrease in the transcript levels of all the three DNMTs in both of the cell lines (Figure 4). The most significant reduction was observed in treatment with decitabine on DNMT1, as compared to all other polyphenols tested in the study.
To further evaluate the effects of the tested natural compounds on the expression of epigenetic regulators, the expression of DNMT1, MeCP2, MBD2, HDAC1, and p21WAF1 proteins were determined in MCF 7 and MDA MB 231 cells by Western blotting. The 2- to 3-folds decrease in the levels of DNMT1 and MeCP2 were observed in the MCF7 and MDA MB 231 cells. Down regulation was also observed in the expression of HDAC1 in the MDA MB 231 cells. However, no effect was observed on the expression levels of MBD2 in both cell lines (Figure 5). In addition to these proteins, the treatment with all the compounds increased the level of p21WAF1 in both the cell lines.
Epigenetic gene regulation has been recognized to play a crucial role in the etiology of cancer. DNA methylation and post translational histone modifications are important epigenetic events in the regulation of gene expression and maintenance of cellular function which may contribute to cancer development [23]. Abnormal methylation in DNA is a hallmark of cancer and often leads to silencing of the tumor suppressor genes, which leads to cancer development and progression. Dietary phytochemicals have been shown to be involved in the epigenetic modifications to regulate the cellular functions and to modify the risks of cancer [4-7,10,24].
In this study, we investigated whether the treatment of breast cancer cells with natural compounds, viz., EGCG, genistein, curcumin, resveratrol, withaferin A, and guggulsterone, resulted in a reversal of the epigenetic changes. We used decitabine, a well characterized DNA methyltransferase inhibitor, as control [4]. Our study reveals that EGCG completely demethylated RARβ2 and TMS1 in the MDA MB 231 cells and RARβ2 in the MCF 7 cells. Genistein treated MDA MB 231 cells showed complete demethylation of Cyclin D2. However, partial demethylation was observed in Cyclin D2 and MGMT on treatment with EGCG; and MGMT was partially demethylated on treatment with genistein. We also observed that the treatment of breast cancer cells at the lower concentrations of curcumin, resveratrol, withaferin A, and guggulsterone for longer time period (more than 96 hours) did not change the methylation status of our panel of genes (data not shown). All the natural compounds tested here downregulated the expression of DNMT1 in both of the breast cancer cell lines.
Histone modifications are epigenetic marks linked to the transcriptional activators and repression of genes [25]. Post-transcriptional modification of histone acetylation/methylation may contribute to cancer development by modulation of the expression of tumor suppressor genes and oncogenes. HDAC plays a key role in the regulation of histone deacetylation. Our data demonstrate that all natural compounds tested, viz., EGCG, genistein, curcumin, resveratrol, withaferin A, and guggulsterone, significantly decreased the HDAC1 expression.
Promoter hypermethylation mediated enhancement of the epigenetic gene silencing involves the activity of MBD proteins, which specifically bind to methylated DNA [26]. The physical association of MBDs with methylated DNA causes steric hindrance for transcription factor binding. Studies demonstrate that at least 2 members of the MBD family, viz., MBD2 and MeCP2 are expressed in the human breast cancer cells and may be involved in gene repression. MeCP2 is able to bind to a single symmetrically methylated CpG [21,22]. It interacts with a transcriptional repressor complex containing HDACs (histone deacetylases) and the transcriptional corepressor Sin3a [26]. Hence, the protein expression of MeCP2 and MBD2 was also evaluated after treatment with EGCG, genistein, curcumin, resveratrol, withaferin A, and guggulsterone. The expression of MeCP2 was reduced by 2 to 3 folds. However, no effect was observed on the expression levels of MBD2.
In the cancer cells, the epigenetic changes and the coexisting genetic lesions often fundamentally cripple various biochemical pathways, e.g., cell cycle arrest and apoptosis. DNMT1 and p21WAF1 compete for the same binding site on PCNA; and an increase in the DNMT1 expression maypromote the dissociation of p21WAF1 from PCNA, perhaps making p21WAF1 more susceptible to ubiquitination and proteasomal degradation [27,28]. A decrease in the DNMT1 expression would then be expected to have an opposite effect on the p21WAF1 stability [27,28]. We also evaluated the effects of these natural compounds on p21WAF1 and observed that the treatments with EGCG, genistein, curcumin, resveratrol, withaferin A, and guggulsterone resulted in 2 to 4 folds increase in the p21WAF1 expression.
In summary, similar to the previous reports, we demonstrated that EGCG and genistein can restore or reactivate the expression of the DNA hypermethylated silenced genes in breast cancer by the down regulation of DNMT1. Recently, the molecular docking of the interactions between curcumin and DNMT1 [8] have suggested that curcumin covalently blocks the catalytic thiolate of DNMT1 to exert its inhibitory effect. Some recent studies have suggested that other polyphenols, lacking a gallic/pyrogallic acid moiety, cannot form a similar strong coordination with the DNMT catalytic center, which, in turn, interferes with the activities of DNMTs inhibition [24]. Hence, we presume that one of the plausible explanations for the curcumin and all other compounds tested not reversing the methylation of some genes may be due to their weak effects on the DNMT catalytic center. Another explanation may be indicating of the functional importance of MBD2. Recent studies on the structure of MBD2 bound to a methylated gene target sequence suggest that the sequence context may direct this preferential binding of MBD2 to certain CpG rich regions [29].
The findings of the current study are preliminary and indicate that the natural compounds, viz., EGCG, genistein, curcumin, resveratrol, withaferin A, and guggulsterone, may provide cancer preventive activity through the modification of the epigenetic process of the gene silencing. However, in-depth future mechanistic studies aimed to elucidate how these compounds affect the gene transcription are warranted. It reinforces the view that these agents can exert inhibitory activity and may be useful in the investigation of the effects of the dietary natural compounds as adjuvants in cancer therapies.
Figures and Tables
 | Figure 1Expression levels of DNMT1, DNMT 3a, and DNMT3b in breast cancer tissues. The levels of DNMT1, DNMT3a, and DNMT3b mRNA were observed to be 1.2- to 4.4-folds, 1.1- to 3.77-folds, and 1.06- to 4.01-folds elevated in most of the breast cancer tissues as compared to the adjacent normal breast tissues. |
 | Figure 2Effect of natural compounds on cell viability of breast cancer cells. The effect of natural compounds, viz., curcumin, EGCG, resveratrol, genistein, and guggulsterone on the viability of breast cancer cells was determined by MTT assay. (A) MCF 7. (B) MDA MB 231. (C) Effect of withaferin A on MCF 7 and MDA MB 231 cells. Mean values from the three experiments±standard error of mean (SEM) are shown. |
 | Figure 3Effect of natural compounds on the methylation status of ERα, TMS1, PRB, Maspin, RARβ2, SLIT2, Cyclin D2, and MGMT. MCF 7 and MDA MB 231 cells were treated with EGCG, genistein, withaferin A, curcumin, resveratrol, guggulsterone (conc. of each natural compound used equals their IC50 value) and decitabine (at conc. of 10 µM for MDA MB 231 cells, and 12 µM for MCF 7 cells) for 96 hours. MSP was performed for ERα, TMS1, PRB, Maspin, Cyclin D2, and MGMT in MDA MB 231 cells and RARβ2 and SLIT2 in MCF 7 cells. |
 | Figure 4Effect of natural compounds on the DNMT transcript levels, viz., DNMT1, DNMT3a, and DNMT3b in human breast cancer cell lines. MCF 7 and MDA MB 231 cells were treated with EGCG, genistein, withaferin A, curcumin, resveratrol, guggulsterone (conc. of each compound used equals their IC50 value) and decitabine (at conc. of 10 µM for MDA MB 231 cells and 12 µM for MCF 7 cells) for 96 hours and then real time polymerase chain reaction analysis was performed for the transcript levels of DNMT1 (A), DNMT3a (B), and DNMT3b (C). |
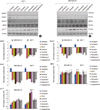 | Figure 5Effect of natural compounds on the expression of p21WAF1, DNMT1, HDAC1, and methyl-CpG binding proteins (MeCP2 and MBD2) in human breast cancer cell lines. MCF 7 and MDA MB 231 cells were treated with EGCG, genistein, withaferin A, curcumin, resveratrol, guggulsterone (conc. of each compound equals their IC50 value) and decitabine (at conc. of 10 µM for MDA MB 231 cells and 12 µM for MCF 7 cells) for 96 hours and then Western blot analysis was performed for DNMT1, HDAC1, MeCP2, MBD2, and p21WAF1 in MCF 7 cells (A) and MDA MB 231 cells (B). The fold change values were calculated as a relative change in comparison to the control cells treated with DMSO (expression equals 1). |
ACKNOWLEDGEMENTS
S.M. is thankful to UGC for providing him with the SRF Fellowship. The authors thank Dr. Fayaz Malik, the Indian Institute of Integrative Medicine (Council of Scientific and Industrial Research), for the kind gift of withaferin A. R.R. gratefully acknowledges the financial support from Canadian Institutes of Health Research for the Chair in the Advanced Cancer Diagnostics.
References
2. Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu Rev Pharmacol Toxicol. 2008. 48:257–276.

3. Siedlecki P, Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim Pol. 2006. 53:245–256.

4. Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005. 97:1498–1506.

5. Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003. 63:7563–7570.
6. Fini L, Selgrad M, Fogliano V, Graziani G, Romano M, Hotchkiss E, et al. Annurca apple polyphenols have potent demethylating activity and can reactivate silenced tumor suppressor genes in colorectal cancer cells. J Nutr. 2007. 137:2622–2628.

7. King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008. 49:36–45.

8. Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009. 19:706–709.

9. Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009. 30:662–670.

10. Ramos S. Effects of dietary flavonoids on apoptotic pathways related to cancer chemoprevention. J Nutr Biochem. 2007. 18:427–442.

11. Choi JS, Choi YJ, Park SH, Kang JS, Kang YH. Flavones mitigate tumor necrosis factor-alpha-induced adhesion molecule upregulation in cultured human endothelial cells: role of nuclear factor-kappa B. J Nutr. 2004. 134:1013–1019.

12. Sim GS, Lee BC, Cho HS, Lee JW, Kim JH, Lee DH, et al. Structure activity relationship of antioxidative property of flavonoids and inhibitory effect on matrix metalloproteinase activity in UVA-irradiated human dermal fibroblast. Arch Pharm Res. 2007. 30:290–298.

13. Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005. 68:1018–1030.

14. Lee WJ, Zhu BT. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006. 27:269–277.

15. Chen D, Milacic V, Chen MS, Wan SB, Lam WH, Huo C, et al. Tea polyphenols, their biological effects and potential molecular targets. Histol Histopathol. 2008. 23:487–496.
16. Chen D, Wang CY, Lambert JD, Ai N, Welsh WJ, Yang CS. Inhibition of human liver catechol-O-methyltransferase by tea catechins and their metabolites: structure-activity relationship and molecular-modeling studies. Biochem Pharmacol. 2005. 69:1523–1531.

17. Moiseeva EP, Almeida GM, Jones GD, Manson MM. Extended treatment with physiologic concentrations of dietary phytochemicals results in altered gene expression, reduced growth, and apoptosis of cancer cells. Mol Cancer Ther. 2007. 6:3071–3079.

18. Mirza S, Sharma G, Prasad CP, Parshad R, Srivastava A, Gupta SD, et al. Promoter hypermethylation of TMS1, BRCA1, ERalpha and PRB in serum and tumor DNA of invasive ductal breast carcinoma patients. Life Sci. 2007. 81:280–287.

19. Sharma G, Mirza S, Prasad CP, Srivastava A, Gupta SD, Ralhan R. Promoter hypermethylation of p16INK4A, p14ARF, CyclinD2 and Slit2 in serum and tumor DNA from breast cancer patients. Life Sci. 2007. 80:1873–1881.

20. Shukla S, Mirza S, Sharma G, Parshad R, Gupta SD, Ralhan R. Detection of RASSF1A and RARbeta hypermethylation in serum DNA from breast cancer patients. Epigenetics. 2006. 1:88–93.

21. Garinis GA, Patrinos GP, Spanakis NE, Menounos PG. DNA hypermethylation: when tumour suppressor genes go silent. Hum Genet. 2002. 111:115–127.

22. Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000. 16:168–174.

24. Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010. 17:2141–2151.

25. Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001. 13:263–273.

26. Prokhortchouk E, Hendrich B. Methyl-CpG binding proteins and cancer: are MeCpGs more important than MBDs? Oncogene. 2002. 21:5394–5399.

27. Fang JY, Lu YY. Effects of histone acetylation and DNA methylation on p21( WAF1) regulation. World J Gastroenterol. 2002. 8:400–405.





 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download