Abstract
Purpose
Hypermethylation of the tumor suppressor genes is frequently observed in the tumor development and progression. However, the correlation between the hypermethylation of the tumor suppressor genes, CDH1 and the axillary lymph node (ALN) metastasis is not fully elucidated. To verify the role of the CDH1 promoter hypermethylation in the ALN metastasis and prognosis, we compared the methylation status of the CDH1 genes in the primary lesion and the paired metastatic ALNs.
Methods
We selected a total of 122 paraffin-embedded specimens of the primary and paired metastatic lymph node from 61 breast cancer patients and analyzed the frequency of hypermethylation in the primary and metastatic lymph node using the methylation-specific polymerase chain reaction. In addition, the methylation status of CDH1 was analyzed with the clinicopathologic characteristics, the disease-free survival and disease-specific survival.
Results
The hypermethylation of CDH1 gene was identified in 54 (88.5%) of the 61 patients who had axillary metastasis. The hypermethylation status of the CDH1 gene was significantly increased in the metastatic ALNs compared with that in the primary tumors (60.7% vs. 45.9%, p<0.001). The hypermethylation status of the CDH1 genes in the metastatic ALNs was associated with a poor histologic grade (p=0.041) and the patients who had methylated tumor in the primary lesion showed worse disease-free survival than the patients who did not have methylated tumor (p=0.046).
Although the advances in the early detection and the treatment modalities of breast cancer have achieved the improved survival, there is still a significant number of mortalities. Breast cancer is the leading contributor of to cancer mortality in women, and there are more than one million new cases per year throughout the world [1]. Cancer metastasis is a crucial factor in the prediction of the disease progression and patient survival, and the axillary lymph node (ALN) is known to be the most frequent metastatic site. the metastasis of ALN is an essential component in determining the tumor stage and the treatment strategies [2].
Cancer development is a result of the complex genetic and epigenetic alterations, and these genetic changes eventually affect the cancer cell progression and metastasis [3]. In the last several decades, a group of genes called the tumor suppressor genes have been researched. These genes are expressed in the normal tissues but not in the cancer tissues, where the tumor suppressor genes are frequently silenced and hypermethylated at the promoter regions in the cancer tissues [4]. In particular, the hypermethylation of the CpG island, where the CpGs are concentrated, leads to an inactivation of these tumor suppressor genes, ultimately leading to the inability to impede malignant transformation [5]. To date, several studies have revealed the hypermethylated and silenced genes at the promoter regions in the breast cancer cells [4]. For example, the mismatch repair genes hMLH1 and hMSH2 are mutated in breast cancer [6]; and the retinoic acid beta 2 (RARβ2) receptor is methylated and silenced in a fraction of breast cancers, which leads to the breast cancer progression [7].
The CDH1 gene encodes for the transmembrane glycoprotein E-cadherin that is involved in the epithelial cell adhesion, and a loss of its expression is associated with the release of invasive tumor cells from the primary tumor [8]. In general, the CDH1 mutation results in a loss of the E-cadherin expression and is detected in a diffuse type gastric cancer and the infiltrative lobular carcinoma of the breast [9,10]. Recent studies have reported that a reduced or absent E-cadherin expression is also found in the invasive ductal carcinoma of the breast [11]. Furthermore, hypermethylation of the CDH1 gene was reported to involve the sentinel lymph node metastasis [12]. However, there are not enough studies on the role of the hypermethylation of the CDH1 gene in the axillary lymph node metastasis and the prognosis for the Korean breast cancer patients.
For this reason, we evaluated the methylation status of the CDH1 genes in the primary and the matched metastatic ALN to evaluate the role of the CDH1 gene in the ALN metastasis. We also analyzed and compared the clinicopathologic characteristics and prognosis of the patients according to the methylation status of CDH1.
A total of 92 patients, who had undergone breast cancer surgery because from the primary breast cancer and a concurrent ALN metastasis between April 2002 and December 2004 in the Korea University Hospital, Seoul, Korea were selected. The selection criteria for this study included 1) diagnosis with invasive ductal carcinoma; 2) ALN metastasis at the same time of surgery or within 1 month after surgery; 3) enough tumor tissues both in the primary lesion and in the metastatic ALN; and 4) without preoperative chemotherapy or radiation therapy. The carcinomas with massive necrosis or the inflammatory carcinoma were also excluded, as these cancer tissues pose difficulties in the cancer cell extraction to assure the suitable sample quality. At last, 61 patients were selected. The study was approved by the Korea University Anam Hospital Institutional Review Board.
All cancer tissues were routinely processed and diagnosed. The histology was confirmed by a pathologist. The estrogen and progesterone receptor status were evaluated in the primary tumor by the standard immunohistochemistry (Dako, Glostrup, Denmark). The tumors were considered positive if more than 1% of the cells showed nuclear staining. HER2 status was determined on the primary tumor tissues with the anti-HER2 monoclonal antibody (Lab Vision, Kalamazoo, USA); and considered positive if the staining intensity score was 3 or a score 2 with a fluorescence in situ hybridization positive. The stage of cancer was classified according to the 7th edition of tumor, node, metastasis system (TNM) by the American Joint Committee on Cancer (AJCC) [2].
The cancer area was marked on the hematoxylin-eosin (H&E) stained slides of the primary tumors and the metastatic ALNs. The corresponding area from 4 to 5 tissue sections of 4-µm thick, formalin-fixed and paraffin-embedded tissue blocks were scraped. The tissue sections were deparaffinized in xylene followed by ethanol incubation. Genomic DNA was isolated using a GENE ALL™ Tissue SV Kit (GeneAll Biotechnology, Seoul, Korea) according to the manufacturer's recommendation. Briefly, the tissue samples were digested with proteinase K, and the DNA samples were bound to columns, washed and eluted. All paraffin-fixed tissues were centrifuged with 1,200-µL xylene and washed with ethanol. After being mixed with 20-µL proteinase K solution, the deparaffinized tissues were incubated at 56℃ for 2 hours. Finally, SV column and buffer were added in the tubes and centrifuged with the tissue samples. Supernatants were used for sodium-bisulfite modification.
Extracted DNA was modified with sodium bisulfite using the EZ DNA Methylation™ Kit (Zymo Research, Orange, USA) following the kit protocols. Purified DNA was denatured with a dilution buffer and incubated with the CT conversion reagent (Zymo research) at 50℃ for 12 to 16 hours. The modified DNA was applied to columns (Zymo-Spin™ IC Column; Zymo Research) and centrifuged with 100-µL wash buffer. After the washing phase, 200-µL desulphonation buffer was added to the column, and the DNA was incubated at room temperature (20-30℃) for 20 minutes. Finally, the substrates were centrifuged at 30 seconds with an elution buffer. In this modification, the unmethylated cytosines were converted to uracils, whereas the methylated cytosines were unaffected in the reaction and remained as cytosines.
The sodium bisulfite-converted DNA was amplified with Blend Taq®Plus (Toyobo, Osaka, Japan), using specific primers in the optimizing annealing temperature (Table 1) [13]. The DNA samples were predenatured at 94℃ for 2 minutes. Subsequently, the polymerase chain reaction (PCR) amplification was accomplished using a 35 timed cycles of denaturation at 94℃ for 30 seconds, annealing for 30 seconds and extension at 72℃ for 1 minute, according to the manufacturer's indication. The postmethylation specific PCR products were loaded on the SeaKem® LE Agarose (Lonza, Rockland, USA) and analyzed using electrophoresis. Figure 1 represents the status of methylation on the agarose gel after the electrophoresis. We used 100 bp DNA Ladder Marker (Enzynomics, Daejeon, Korea) as a molecular weight marker and the breast cancer cell line MDA-MB-231 as a positive marker, which demonstrated the methylation and silencing of the CDH1 [14,15].
Descriptive analyses were performed to explore the clinicopathologic characteristics and the methylation status. Pearson's chi-square test or Fisher's exact test was used for binomial comparison. Logistic regression test was used for the multivariate analysis. We used the Cox hazard regression method to estimate the relapse-free survival and the disease-specific survival according to the methylation status.
Data were analyzed using Microsoft Excel 2007® (Microsoft Corp., Redmond, USA). The statistical analyses were performed using PASW® Statistics version 18.0 (SPSS Inc., Chicago, USA). Reported p-values were two-sided, and the statistical significance was set at p<0.05.
Characteristics of the 61 patients included in this study are presented in Table 2. All patients were female, and the mean age at diagnosis was 47.1 years (range, 30-72 years). According to the AJCC TNM stage [2], T2 (33 patients, 54.1%), N1 (40 patients, 65.6%), and stage IIB (23 patients, 36.1%) were most common in each of the subgroups. Fifty-nine cases were invasive ductal carcinomas, not otherwise specified, and two cases were mixed with tubular carcinoma and mucinous carcinoma. Thirty-two cases (52.5%) were classified as the poorly differentiated histologic grade (G3). The hormone receptor and HER-2 status were assessed in all primary tumors. Thirty-seven cases (60.7%) were estrogen receptor positive, and 33 cases (54.1%) were progesterone receptor positive. HER2 was overexpressed in 25 cases (41.0%) of primary tumors.
The methylated rate of the CDH1 gene in the primary tissues and the metastatic ALNs is described in Table 3. The methylation rate of CDH1 gene was higher in the metastatic node than in the primary site (60.7% vs. 45.9%, p<0.05). In addition, we compared the association of the methylation status between the primary and the matched axillary node (Table 4). Ten cases of the unmethylated primary tumors were methylated in the matched metastatic lymph nodes, while only one case of the methylated primary tumor was unmethylated in the matched lymph node.
The methylated status and the clinicopathologic characteristics were analyzed with the univariate and multivariate methods (Table 5). The analyses contained these factors: patient age, tumor stage, histologic grade, hormone receptor status, HER2/neu overexpression and lymphovascular invasion status. In univariate and multivariate analysis, the methylation status of the CDH1 gene in the primary tumor was not associated with any variables (Table 5). However, CDH1 methylation in the metastatic ALN was correlated with a poor histologic grade in logistic regression analysis (Table 6).
After a median follow-up of 88 months (range, 1-138 months), 18 cases of any local or distant recurrences and 8 cases of disease-specific mortality were occurred. The overall 5-year cumulative relapse-free rate was 76.0%±5.8% and disease-specific survival rate was 85.4%±4.8%. The univariate analysis showed that the methylation status either in the primary lesion or the metastatic ALNs was not correlated with relapse-free survival (RFS) and disease-specific survival (DSS). For patients who had methylated primary tumor, the 5-year cumulative relapse-free rate was 66.7%±9.7%, and the disease-specific survival rate was 83.8%±7.4%, compared with 79.5%±7.6% (p=0.131) and 86.6%±6.2% (p=0.697) in patients who had the nonmethylated primary tumor. Patients who had methylated ALNs also did not show statistical differences for RFS (p=0.895) and DSS (p=0.184) in the univariate analysis.
To exclude the potential confounders, the databases were adjusted for the age, T, N stage, tumor grade, presence of lymphovascular invasion, estrogen receptor expression, use of chemotherapy, and use of hormone therapy using multivariate analysis. Figure 2 describes the adjusted cumulative RFS rate according to the methylated status of the primary tumor and the metastatic lymph node. Hypermethylation of the CDH1 primer in the primary tumor was significantly correlated with poor RFS (p=0.046), while the status of hypermethylation in the metastatic axillary node was not correlated with RFS (p=0.105). The methylation status of the CDH1 gene in the primary and metastatic axillary node had no significant correlations with of DSS (p=0.911 and p=0.906, respectively).
Although breast cancer is a highly metastatic disease, prognosis may be milder if it is diagnosed prior to the metastasis. ALN is the most common and the first metastatic site, and the status of the ALN plays an important role in the prediction of breast cancer prognosis [16]. To date, there has been a lack of satisfying tools in preoperatively identifying the ALN metastasis in the early breast cancer patients. ALN dissection and sentinel lymph node (SLN) biopsy are invaluable methods in recognizing the presence of the tumor cells [17,18]. In this study, the metastatic status of ALN was recognized with a SLN biopsy (18 cases) and initial ALN dissection (43 cases). Because SLN biopsy and ALN dissection may cause some complications including pain, numbness, lymphedema and a limitation of the arm movement [19], an alternative less invasive method to detect the ALN metastasis is necessary.
The methylation of several tumor suppressor genes is a well-known mechanism of the oncogenesis in breast cancer [3,6,7]. Identifying the methylation pattern of the specific genes related with the axillary lymph node metastasis may be a crucial key to understanding the disease progression and metastasis [12,20]. In this study, we focused on the relationship between the methylation of the specific tumor suppressor gene and the ALN metastasis. The hypothesis of this study was that if the specific tumor suppressor genes were related with the tumor metastasis, the hypermethylation of these genes would be identified in the metastatic axillary lymph node in an equal or higher frequency as in the primary lesion [20].
CDH1 is known as a E-cadherin gene and located on chromosome 16q22.1. Recent studies have reported that a loss of E-cadherin expression is associated with the nodal metastasis [21] and distant metastasis including those in the bone, brain, and lungs [22]. Hypothesizing on the hypermethylation frequency of the tumor suppressor genes in the metastatic axillary lymph node, our results have shown that the methylation of CDH1 is detected more frequently in the metastatic ALN than in primary tumor. Comparing the primary tumor and its matched ALN, CDH1 was more frequently methylated in the matched metastatic lesion. Based on these results, we suggest that CDH1 may be involved in the ALN metastasis.
Some studies have reported that the reduced E-cadherin expression may be associated with the poor prognostic factors, such as a high histologic grade, nodal metastasis and a loss
of hormone receptor expression [23,24]. On the other hand, Singhai et al. [25] reported that a loss of E-cadherin expression did not show a correlation with the prognostic variables. So far, there has been a debate on the relationship between the E-cadherin expression and prognosis. In this study, we found that the CDH1 methylation in the metastatic ALN was correlated with a high histologic grade in the multivariate analysis. Furthermore, we also demonstrated that the hypermethylation of CDH1 in the primary lesion was significantly correlated with the RFS.
In general, the HER2 positive breast cancer patients compose approximately 15% to 20% of all breast cancers cases [26]. The HER2 positive rate is reported to increase up to 40% in the aggressive tumor characters and the high-stage disease [27]. In this study, the patient demographic showed a high rate of HER2 receptor expression (41.0%). More than half of the cases were of the poorly differentiated histologic grade (G3), and all the carcinomas had axillary metastasis. The high rate of HER2 expression in this study may arise from the aggressive study cohort. The expression of the HER2 receptor is a well known prognostic factor in breast cancer [28]. Hence, there may be a question of the high rate of HER2 expression affecting the worse prognosis of patients with the CDH1 methylated tumor. We performed the multivariate analysis to exclude the potential confounders including HER2 expression, and our analysis showed that the HER2 expression did not affect the disease progression (p=0.437 in the RFS, and p=0.411 in the DSS) in this study cohort.
One of the limitations in this study was that we did not show the relation between the E-cadherin expression level and the CDH1 methylation status. While we planned to analyze the E-cadherin expression level according to the CDH1 methylation status, the correlation of the CDH1 methylation and the E-cadherin expression have been well-established in several studies [29,30]. As such, we focused on the effects of the CDH1 hypermethylation to the axillary metastasis and prognosis.
In conclusion, this study demonstrated that the methylation status of the tumor suppressor genes was different in the primary breast cancer and the matched metastatic axillary lymph node; and that the methylation rate of CDH1 was higher in the metastatic node than the primary site. We suggest that the hypermethylation of CDH1 may be extended from the primary tumor to the axillary metastatic node during the tumor progression. In addition, the methylation status of CDH1 was associated with a poor RFS in both the primary tumor and the metastatic axillary lymph node. Taken together, we suggest that the methylation status of the CDH1 gene may be a promising biomarker for assessing the disease status in the breast cancer.
Figures and Tables
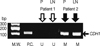 | Figure 1Representative methylation-specific polymerase chain reaction results for the CDH1 genes. Note the unmethylated primary tumor and the metastatic axillary lymph node in patient 1 (hollow arrows). Solid arrows (patient 2) indicate the methylated primary tumor and the metastatic axillary lymph node.
P=primary tumor; LN=axillary lymph node; M.W.=molecular weight (bp=base pair); P.C.=positive control; M=methylated tissue; U=unmethylated tissue.
|
 | Figure 2Multivariate analysis curves for the relapse-free survival (RFS) according to the methylation status. (A) RFS according to the methylation status of the primary tumor (p=0.046). (B) RFS according to the methylation status of the metastatic axillary node (p=0.352).
Primary=methylation status in primary tumor; LN=methylation status in axillary lymph node.
|
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011. 61:69–90.

2. Edge SB. American Joint Committee on Cancer. AJCC Cancer Staging Manual. 2010. New York: Springer;648.
3. Hunter KW, Alsarraj J. Gene expression profiles and breast cancer metastasis: a genetic perspective. Clin Exp Metastasis. 2009. 26:497–503.

4. Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996. 93:9821–9826.

5. Hayslip J, Montero A. Tumor suppressor gene methylation in follicular lymphoma: a comprehensive review. Mol Cancer. 2006. 5:44.
6. Murata H, Khattar NH, Kang Y, Gu L, Li GM. Genetic and epigenetic modification of mismatch repair genes hMSH2 and hMLH1 in sporadic breast cancer with microsatellite instability. Oncogene. 2002. 21:5696–5703.

7. Fackler MJ, McVeigh M, Evron E, Garrett E, Mehrotra J, Polyak K, et al. DNA methylation of RASSF1A, HIN-1, RAR-beta, cyclin D2 and twist in in situ and invasive lobular breast carcinoma. Int J Cancer. 2003. 107:970–975.

8. Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004. 1014:155–163.

9. Guilford PJ, Hopkins JB, Grady WM, Markowitz SD, Willis J, Lynch H, et al. E-cadherin germline mutations define an inherited cancer syndrome dominated by diffuse gastric cancer. Hum Mutat. 1999. 14:249–255.

10. Berx G, Cleton-Jansen AM, Strumane K, de Leeuw WJ, Nollet F, van Roy F, et al. E-cadherin is inactivated in a majority of invasive human lobular breast cancers by truncation mutations throughout its extracellular domain. Oncogene. 1996. 13:1919–1925.
11. Szyf M, Pakneshan P, Rabbani SA. DNA methylation and breast cancer. Biochem Pharmacol. 2004. 68:1187–1197.

12. Shinozaki M, Hoon DS, Giuliano AE, Hansen NM, Wang HJ, Turner R, et al. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res. 2005. 11:2156–2162.

13. Righini CA, de Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007. 13:1179–1185.

14. Corn PG, Heath EI, Heitmiller R, Fogt F, Forastiere AA, Herman JG, et al. Frequent hypermethylation of the 5' CpG island of E-cadherin in esophageal adenocarcinoma. Clin Cancer Res. 2001. 7:2765–2769.
15. Gagnon J, Shaker S, Primeau M, Hurtubise A, Momparler RL. Interaction of 5-aza-2'-deoxycytidine and depsipeptide on antineoplastic activity and activation of 14-3-3sigma, E-cadherin and tissue inhibitor of metalloproteinase 3 expression in human breast carcinoma cells. Anticancer Drugs. 2003. 14:193–202.

16. Fitzgibbons PL, Page DL, Weaver D, Thor AD, Allred DC, Clark GM, et al. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000. 124:966–978.
17. Veronesi U, Paganelli G, Viale G, Luini A, Zurrida S, Galimberti V, et al. A randomized comparison of sentinel-node biopsy with routine axillary dissection in breast cancer. N Engl J Med. 2003. 349:546–553.

18. Giuliano AE, Haigh PI, Brennan MB, Hansen NM, Kelley MC, Ye W, et al. Prospective observational study of sentinel lymphadenectomy without further axillary dissection in patients with sentinel node-negative breast cancer. J Clin Oncol. 2000. 18:2553–2559.

19. Warmuth MA, Bowen G, Prosnitz LR, Chu L, Broadwater G, Peterson B, et al. Complications of axillary lymph node dissection for carcinoma of the breast: a report based on a patient survey. Cancer. 1998. 83:1362–1368.

20. Feng W, Orlandi R, Zhao N, Carcangiu ML, Tagliabue E, Xu J, et al. Tumor suppressor genes are frequently methylated in lymph node metastases of breast cancers. BMC Cancer. 2010. 10:378.

21. Hunt NC, Douglas-Jones AG, Jasani B, Morgan JM, Pignatelli M. Loss of E-cadherin expression associated with lymph node metastases in small breast carcinomas. Virchows Arch. 1997. 430:285–289.

22. Mbalaviele G, Dunstan CR, Sasaki A, Williams PJ, Mundy GR, Yoneda T. E-cadherin expression in human breast cancer cells suppresses the development of osteolytic bone metastases in an experimental metastasis model. Cancer Res. 1996. 56:4063–4070.
23. Heimann R, Lan F, McBride R, Hellman S. Separating favorable from unfavorable prognostic markers in breast cancer: the role of E-cadherin. Cancer Res. 2000. 60:298–304.
24. Guriec N, Marcellin L, Gairard B, Caldéroli H, Wilk A, Renaud R, et al. E-cadherin mRNA expression in breast carcinomas correlates with overall and disease-free survival. Invasion Metastasis. 1996. 16:19–26.
25. Singhai R, Patil VW, Jaiswal SR, Patil SD, Tayade MB, Patil AV. E-Cadherin as a diagnostic biomarker in breast cancer. N Am J Med Sci. 2011. 3:227–233.

26. Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006. 295:2492–2502.

27. Telli ML, Chang ET, Kurian AW, Keegan TH, McClure LA, Lichtensztajn D, et al. Asian ethnicity and breast cancer subtypes: a study from the California Cancer Registry. Breast Cancer Res Treat. 2011. 127:471–478.

28. Ross JS, Fletcher JA, Linette GP, Stec J, Clark E, Ayers M, et al. The Her-2/neu gene and protein in breast cancer 2003: biomarker and target of therapy. Oncologist. 2003. 8:307–325.

29. Berx G, Van Roy F. The E-cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 2001. 3:289–293.

30. Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995. 55:5195–5199.




 PDF
PDF ePub
ePub Citation
Citation Print
Print








 XML Download
XML Download