Abstract
Purpose
Triple-negative breast cancer, has a significant clinical relevance being associated with a shorter median time to relapse and death and does not respond to endocrine therapy or other available targeted agents. For this reason, identifying the molecular pathways associated with increased aggressiveness, for example the presence of stem cell populations within the tumor and alteration of genes associated with cell cycle regulation represents an important objective in the clinical research into this neoplasm.
Methods
To investigate the role of cell cycle progression inhibitor Geminin in triple-negative breast cancers and its potential correlation with stem-like phenotype of this neoplasm, we used tissue microarray technology to build a specific triple-negative breast cancer tissue micro-array. Geminin and cancer stem cell marker CD133 expression was further investigated at the mRNA level for selected breast tumor samples through realtime polymerase chain reaction quantification.
Results
Our results showed that CD133 expression was significantly associated to high Geminin expression (p=0.017), a strong association between Ki-67 and tumor grade (p=0.020) and an inverse association between Geminin expression and lymphonode metastases (p=0.058), and a trend of statistically significance between Geminin marker expression and survival of triple-negative breast cancer patients (p=0.076).
Breast cancer represents a heterogeneous group of tumors with differences in morphology, in biology and in treatment.
Recent DNA-microarray-expression-profiling studies have classified breast cancers into 5 major subtypes: the luminal A and B subgroups, exhibiting markers for the luminal epithelial layer of normal breast duct with estrogen receptor (ER) positivity; the HER2+ subgroup, overexpressing genes that are co-amplified with ERBB2 (encoding HER2); the basal-like (basaloid) subgroup, sharing markers for myoepithelial layer of normal breast ducts, and the normal breast-like subgroup, exhibiting similar expression patterns of normal breast [1,2]. Basal-like tumors are typically negative for ER, for the progesterone receptor (PR) and for the HER2 but positive for basal cytokeratins (CK5/6/14/17), for epidermal growth factor receptor (EGFR) and/or for c-kit [3].
The triple-negative breast tumor subtypes was define on the basis of an immunohistochemistry and for typically being negative for ER, PR, HER2 represents approximately 20% of all breast carcinomas, however it does not have a definition provided in the World Health Organization (WHO) classification of breast tumors [4].
The molecular subgroup Basal-like represents the majority of triple-negative carcinomas (50-80%) [5]. It is important to note that, whereas triple-negative cancers are frequently found to be basal-like tumors, triple-negative cancers can less commonly fall into any of the other intrinsic subtypes [6].
More recently, the basal-like subtype was found to be associated significantly with mutations in BRCA1, which has been proposed to be a possible stem cell regulator in the breast [7]. Moreover, an association has been demonstrated between the CD44+/CD24- stem-like phenotype to basal-like and BRCA1 hereditary breast cancer [8], and an increased expression of other putative stem cell markers, BMI-1, EZH2, and Oct-4 only in basal-like breast cancers [9]. These findings support the hypothesis that basal-like cancers might be related to a more undifferentiated stem cell-like lineage [10].
Recent concepts in cancer development suggest that a minority population of cancer stem-like cells may determine the biological behavior of tumors, including response to therapy [11]. Although the cell of origin of basal breast cancer has not been identified conclusively, a link to the progenitor/stem cell compartment of the mammary epithelium has been proposed [12]. In this context, cell-cycle gene pathways that control the interplay between replication, cell proliferation, survival and differentiation are ideal candidates for stem cell-derived mammary tumorigenesis.
Geminin is a 25-kDa nuclear protein that functions by inhibiting DNA replication. Its activity is achieved by regulating Cdt1 and inducing the formation of pre-replicative complexes by loading mini-chromosome maintenance proteins (Mcm) onto chromatin and limiting DNA replication to once per cell cycle. Several studies have suggested that Geminin expression is a marker of the S/G2/M phase of the cell cycle [13].
Moreover, previous reports have demonstrated that Geminin is frequently overexpressed, in vivo, in a variety of human tumors (kidney, colon, breast, lung cancer and lymphoma) and its expression rises with increasing tumor grade, which correlates to a poor prognosis [14-18].
Recently, studies have proven that in vitro siRNA suppression of Geminin is able to arrest proliferation only of cancer cells by inducing DNA re-replication and DNA damage that spontaneously trigger apoptosis [19].
In this paper we have report on an investigation of Geminin expression in triple-negative breast cancers and we demonstate a strong association between its expression and the presence of cancer stem cell population, identified by CD133/Prominin 1 expression. Moreover, we have verified the prognostic role of CD133 and Geminin in triple-negative breast cancers progression. The study was conducted by immunohistochemistry on a specific tissue microarray (TMA) and that verified the alteration of 2 markers gene expression by using real-time polymerase chain reaction (RT-PCR) quantification.
From 2003 to 2009, 204 patients who underwent a mastectomy, quadrantectomy or metastectomy at the National Cancer Institute "Giovanni Pascale" of Naples, Italy, were enrolled into this study. The study was approved by the Internal Review Board of of the INT Fondazione Pascale (Naples, Italy) (CEI 556/10 of 12/3/2010).
In our institution, the percentage of tumors classified as triple-negative is approximately 15% to 19% of the total number of breast cancer surgeries. All cases of triple-negative and non-triple-negative breast samples were reviewed according to WHO classification criteria, using standard tissue sections and appropriate immunohistochemical slides.
Medical records for all cases of triple-negative and non-triple-negative breast samples were reviewed for clinical information, including histologic parameters that were determined from the H&E slides. The following clinical and pathological parameters were evaluated for each tumor included in the study: patient age at initial diagnosis; tumor size; histologic subtype; histologic grade; nuclear grade; nodal status; number of positive lymph nodes; tumor stage; tumor recurrence or distant metastasis; and type of surgery (for tumor removal).
In addition, all specimens were chacterizated for all routine diagnostic immunophenotypic parameters.
One hundred fifty-nine patients were used for a TMA building, using the most representative areas from each single case. All tumors and controls were reviewed by 2 experienced pathologists (M.D.B., G.B.). If discrepancies occurred between 2 pathologists that reviewed the same case, the discrepancy was resolved through joint analysis of the case. Tissue cylinders with a diameter of 0.6 mm were punched from morphologically representative tissue areas of each 'donor' tissue block and brought into one recipient paraffin block (3×2.5 cm) using a semiautomated tissue array (Galileo TMA).
Immunohistochemical staining was performed on slides from formalin-fixed, paraffin embedded tissues, corresponding to triple-negative TMA and 47 non-triple-negative cases to evaluate the expression of CD133, ER, PR, c-erbB-2, Ki-67, and Geminin markers. Then, paraffin slides were deparaffinized in xylene and rehydrated through graded alcohols. Antigen retrieval was performed with slides heated in 0.01 M citrate buffer (pH 6.0 for CD133, Geminin, PR, c-erbB-2, Ki-67) or Tris-EDTA (pH 9 for ER) in a bath for 20 minutes at 97℃. After antigen retrieval, the slides allow to cool. The slides were rinsed with TBS and the endogenous peroxidase was inactivated with 3% hydrogen peroxide. After protein block (BSA 5% in PBS 1x), the slides were incubated with primary antibody to human CD133 (CD133/1 [AC 133] pure human, dilution 1:150; Myltenyi Biotec, Bergisch Gladbach, Germany) for 1 hour, and to human ERα (Monoclonal Mouse Anti-Human ERα Clone ID5, dilution 1:35; DAKO, Ely, UK), PR (Monoclonal Mouse Anti-Human PR Clone 636, dilution 1:50; DAKO), c-erbB-2 (Polyclonal Rabbit Anti-Human Oncoprotein, dilution 1:300; DAKO), Ki-67 (Monoclonal Mouse Anti-Human Ki-67 Ag Clone MIB-1, dilution 1:75; DAKO), and Geminin (Rabbit polyclonal ab12147-50, dilution 1:400; Abcam, Cambridge, UK) over night. The sections were rinsed in TBS and incubated for 20 minutes with Novocastra Biotinylated Secondary Antibody (RE7103), a biotin-conjugated secondary antibody formulation that recognized mouse and rabbit immunoglobulins. Then the sections were rinsed in TBS and incubated for 20 minutes with Novocastra Streptavidin-HRP (RE7104) and then peroxidase reactivity was visualized using a 3,3'-diaminobenzidine (DAB). Finally, the sections were counterstained with hematoxylin and mounted. Results are interpreted using a light microscope.
Antigen expression was evaluated independently by a pathologist using light microscopy. The pathologist was unaware of the clinical outcome. For each sample, at least 5 fields (inside the tumor and in the area exhibiting tumor invasion, 400) and >500 cells were analyzed. Using a semiquantitative scoring system microscopically and referring to each antigen scoring method in other studies, an observer evaluated the intensity, extent and subcellular distribution of CD133, Geminin, ER, PR, c-erbB-2, and Ki-67.
The cutoff used to distinguish "positive" from "negative" cases was ≥1% ER/PR positive tumor cells. Immunohistochemical analyses of c-erbB-2 expression describe the intensity and staining pattern of tumor cells. Only membrane staining intensity and pattern were evaluated using the 0 to 3+ score as illustrated in the HercepTest kit scoring guidelines. The FDA-recognized test, the Herceptest™ (DAKO), describes 4 categories: no staining, or weak staining in fewer than 10% of the tumor cells (0); weak staining in part of the membrane in more than 10% of the tumor cells (1+); complete staining of the membrane with weak or moderate intensity in more than 10% of the neoplastic cells (2+); and strong staining in more than 10% (3+). Scores of 0 or 1+ were considered negative for HER2/neu expression, 2+ was uncertain, and 3+ was positive. Cases with score 2+ underwent fluorescence in situ hybridization analysis. For the proliferative index Ki-67 was defined as the percentage of immunoreactive tumour cells out of the total number of cells. The percentage of positive cells per case was scored according to 2 different groups: group 1: <5% (low proliferative activity); and group 2: >15% (high proliferative activity).
In scoring CD133 protein expression, both the immunopositivity in the cell membrane and cytoplasm were considered, while for Geminin protein cytoplasm was considered.
The Geminin immunostaining score was defined as high when there were positive cells percentages about 30% to 80%, and low when there was a positive cells percentage <30%. For CD133 immunostaining all values were expressed only in the percentage of positive cells. The expression was defined high when the positive cells percentage was about 8% to 30%, and low when the percentage of positive cells was <8%.
The association between CD133 and Geminin expression with other clinicopathological parameters was conducted using the χ2 and Student's t-test.
The Pearson χ2 test was used to determine whether there was a relationship between the variables included in this study. The level of significance was defined as p value <0.05. Overall survival (OS) and failure-free survival (FFS) curves were calculated using the Kaplan-Meier method. Statistical significance of the association between individual variables and OS and FFS was determined using the log-rank test. All the statistical analyses were carried out using the SPSS version 8.0 software (SPSS Inc., Chicago, USA).
OS was defined as the time from diagnosis (first biopsy) to death by any cause or until the most recent follow-up. FFS was measured as the time from diagnosis to the occurrence of progression, relapse after complete remission, or death from any cause. FFS had a value of zero for patients who did not achieve complete remission. There was a 5-year follow-up period.
Total RNA was isolated from frozen biopsies, from our Institutional Bio-Bank, using RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer's instructions. Samples were treated with RNase-free DNase (Qiagen GmbH) to prevent amplification of genomic DNA. A total of 1 µg RNA was subjected to cDNA synthesis for 1 hour at 37℃ using the Ready To Go You-Primer First-Strand Beads kit (cod. 27-9264-01; Amersham Biosciences Europe Gmbh, Freiburg, Germany) in a reaction mixture containing 0.5 µg random hexamers (GeneAmp RNA PCR Random Hexamers Set N808-0127; Applied Biosystems, Foster City, USA).
Quantitative RT-PCR was performed in a LightCycler system (Roche Molecular Biochemicals, Mannheim, Germany) using TaqMan® analysis. In this system, all reactions were run in glass capillaries with The LightCycler TaqMan Master Mix (cod. 04735536001; Roche Molecular Biochemicals),10 µL, in a volume of 20 µL containing 2 µL of cDNA and 1 µL of specific TaqMan Gene Expression Assays for human CD133 (RealTime Designer Assay cod. 05583055001; Roche Molecular Biochemicals), and Geminin (RealTime Designer Assay cod. 05532957001; Roche Molecular Biochemicals) according to the manufacturer's directions. All reactions were performed in triplicate. The thermal cycling conditions for CD133 included a step of 20 seconds at 95℃ followed by a 40 cycles of 95℃ for 1 second and 60℃ for 20 seconds, while for Geminin included a step of 20 seconds at 95℃ followed by a 35 cycles of 95℃ for 1 second and 58℃ for 20 seconds. The comparative Ct method was employed to determine the human CD133 and Geminin genes variation, using as reference gene TaqMan Endogenous Controls Human ACTB (β-actin) Endogenous Control (RealTime Designer Assay cod. 05532957001; Roche Molecular Biochemicals). We identified a calibrator cell line that represents the unitary amount of the target of interest, consequently the samples express n-fold mRNA relative to the calibrator. Final amounts of target were determined as follows: target amount=2-Ct, where Ct=[Ct (CD133/Geminin)-Ct (ACTB)]sample-[Ct (CD133/Geminin)-Ct (ACTB)]calibrator.
In our casuistry, we have included 204 samples of breast cancers of which 157 were triple-negative (10 lobular, 4 ductal-lobular, 9 medullary, 1 mucinous, 125 invasive ductal breast carcinomas, and 8 triple-negative metastases) and 47 were non-triple-negative samples (6 HER2 positive, 11 ER and PR positive, 1 ER positive, 1 PR positive, 28 ER, PR, and HER2 positive).
The age of patients with triple-negative breast cancer ranged from 24 to 93 years, with an average age of 57 years. Tumors larger than 2 cm were present in 52% (77/148) of patients and metastatic lymphonodes were found in 41.2% (61/148) of patients at operation. Here are the percentages of tumor gradings: 88.5% (131/148) were grade 3; 10.1% (15/148) were grade 2; 2% (3/148) were grade 1. The expression of proliferation factor Ki-67 was high (>20%) in 122/149 cases, and low (≤20%) in 27/149 cases.
The age of the non-triple-negative breast cancer patients ranged 32 to 80 years, with an average of 56 years. Tumors larger than 2 cm were present in 8.51% (4/47) of patients and metastatic lymph nodes were found in 91.5% (43/47) of patients at operation. 76.6% (36/47) of tumors were graded as grade 3, 23.4% (11/47) as grade 2, 0% (0/47) as grade 1. The expression of proliferation factor Ki-67 was high (>20%) in 26/47 cases, and low (≤20%) in 21/47 cases.
CD133 expression was detected by immunohistochemistry on breast samples included in TMA and on 47 non-triple-negative breast cancers, not included in TMA.
CD133 protein expression was detected, excluding the samples that could not be assessed, in 34/159 samples, where the cytoplasm appeared membranous at times. In 17 samples there was a low CD133 expression, while in 17 specimens there was 15% to 30% CD133 expression. In 2 cases, CD133 was expressed at high levels, at about 30% (Table 1, Figure 1). In normal breast and in triple-negative metastases CD133 was absent. In 3 cases of non-triple-negative breast cancers, there was a low CD133 expression (Tables 1, 2).
Geminin expression was detected by immunohistochemistry on breast samples included in TMA and on 47 samples not included in TMA.
Geminin protein was present in most of triple-negative breast samples (142/159), which excluded those samples that could not be assessed. In 69/159 samples, there was a high level of expression (Table 1, Figure 2). In 73/159 samples, in normal breast and in 2 metastases, the Geminin protein expression was very low.
Moreover, in non-triple-negative breast samples, Geminin protein was present at high level of expression only in 16 non-triple-negative breast cancers, while it was shown low or lack of expression in the remaining samples (Table 2).
Based on statistical elaboration of CD133 and Geminin protein expression analysis with the other clinicopathological parameters in triple-negative breast cancers it was shown that CD133 expression was significantly associated with high Geminin expression (p=0.017) (Table 3).
Moreover, we detected a strong association between Ki-67 and tumor grade (p=0.02) and an inverse association between Geminin expression and lymph node metastases (p=0.058) (Table 1). On the contrary there was no significant association of 2 markers with other clinicopathological parameters (Table 1) and Ki-67 expression (Table 3).
On the contrary we do not have a highlighted statistical correlation between the expression of CD133 and Geminin in non-triple-negative breast cancer specimens (p=0.277) (Table 4).
CD133 and Geminin genes expression was evaluated on normal breast, 3 HER2+ and 11 triple-negative samples selected from TMA, by RT-PCR quantification.
In the normal sample, the Geminin expression was active at a low level (between 1- and 3-fold increase) (Figure 3), while CD133 expression was absent (Figure 4). In HER2+ samples it was shown that Geminin (Figure 3) and CD133 were silent (Figure 4), except for one case where it was present at a low expression of CD133. In all triple-negative samples, there was observed a significant increase in Geminin mRNA expression (between a 10- and 100-fold increase) (Figure 3). Moreover, CD133 mRNA expression appeared at intermediate level in all selected triple-negative specimens (between 3- and 10-fold increase) (Figure 4).
The comparison between the levels of gene and protein expression of CD133 and Geminin showed a strong overlap between the 2 methods. In the Table 5, the comparison of expressions on the 15 samples selected for molecular analysis is shown.
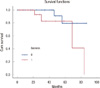
We calculated the OS and FFS for all triple-negative breast cancer samples included in the TMA. We do not have highlighted statistical significance between the expression of CD133 and the FFS (p=0.208) and the OS (p=0.114), and between the expression of Geminin and FFS (p=0.969). Instead we found a trend of statistically significace between the expression of Geminin and OS (p=0.076) (Figure 5).
In this paper we analyzed the expression of a novel proliferation marker, Geminin, and cancer stem cell marker CD133, in a casuistry of triple-negative breast cancers. The analysis was conducted through immunohistochemistry by TMA technology and supported by RT-PCR quantification on selected samples showing a strong correlation between the 2 markers.
Triple-negative breast cancers have a considerable clinical relevance as these types of breast cancer primarily affect young women, appear resistant to conventional chemotherapy regimens, have a particularly poor prognosis, and have a significantly worse clinical outcome than other tumor types [5].
It has been suggested that Geminin, which regulate cell-cycle initiation by impeding a second pre-replication complex assembly once replication had started, may be altered in cancer cells as it has been found to be overexpressed in several human tumors [14-18]. In particular, it has been shown that in several human cancers the high proliferation state is correlated strongly with concomitant high levels of Geminin and Ki-67 [20,21]. In breast cancer Geminin overexpression stimulated cell cycle progression and proliferation in both normal cells and cancer cells and increased the anchorage independent growth of breast cancer cells [22]. Moreover, it was found that increased Geminin expression is a powerful independent indicator of adverse prognosis in invasive breast cancer [14].
More recently, Geminin was inserted in a panel of genes correlated to defective homologous recombination (HR), a DNA double-strand break repair system, frequent in triple-negative breast cancer, and present in a subset of other subtypes, identifying breast cancers that may benefit from therapies that target defective HR such as PARP inhibitors [23].
The hypothesis that mammary carcinogenesis results from the deregulation of normal stem cell self-renewal pathways suggests that components of these pathways might provide attractive targets for therapeutic development [24]. This idea is supported by recent description that the residual breast tumor cell populations surviving after conventional treatment may be enriched for subpopulations of cells with self-renewal features [25].
CD133 is a 5-trans-membrane cell surface glycoprotein located in plasma membrane protrusions where it could act as a regulator of lipid composition, cell polarity and migration. Cancer stem cells in specific tumor types are associated with elevated expression of the stem cell surface marker CD133 [26]. Given the strong rationale linking CD133 expression to more aggressive cellular behavior, including resistance to chemotherapy and radiotherapy, several studies have addressed its correlation with clinicopathological characteristics of cancer patients: in particular, a direct correlation between CD133 expression and advanced stage of disease as well as a poor grade of differentiation has been shown in hepatocellular carcinoma, colon, and malignant gliomas [27]. Recently, the detection of CD133 expression has been reported in invasive ductal breast carcinomas [28] and it was shown, that the expression of CD133 protein could be correlated with tumor size, metastasis to axillary lymph nodes and clinical stage of disease [29].
In this paper, we used the CD133 marker to identify cancer stem cells population in triple-negative breast cancers and related with clinicopathological parameters, Geminin expression and patients survival.
Our results showed a strong overlap between gene and protein expression of these markers in triple-negative breast cancer specimens. Moreover, we showed a consistent correlation between Geminin and CD133 expression in triple-negative samples suggesting the possibility that the cell cycle pathway linked to Geminin activity can be associated to preservation and propagation of cancer stem cells.
On the contrary, no significant associations exist between Geminin and Ki-67 markers, as reported in other studies.
Finally, we correlated the immunohistochemistry expression data for both CD133 and Geminin with triple-negative breast cancers patients survival to establish their potential role in the tumor evolution and progression, showing an inverse trend of statistical significance between Geminin expression and survival, while we haven't found a statistical significance with the expression of CD133.
Numerous studies of gene profiling have attempted to clarify the correct definition of the molecular profile of the triple-negative breast cancers, in order to establish better stratification of patients for therapeutic purposes.
As it is not entirely known which molecular pathways may be linked to the presence of cancer stem cells, we found that the strong association between the expression of CD133 and Geminin, could be represent a piece to include in the complex puzzle that characterizes molecular profiles of breast tumors and in particular of triple-negative breast cancers.
Figures and Tables
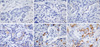 | Figure 1CD133 immunostaining. (A) Immunonegativity for CD133 in normal breast cells (×60). (B) Immunonegativity for CD133 in HER2+ breast sample (×60). (C) CD133 low cytoplasmatic expression in triple-negative (TN) sample 27 (×60). (D) CD133 moderate cytoplasmatic expression in TN sample 131 (×60). (E) CD133 high cytoplasmatic expression in TN sample 7 (×60). (F) CD133 high membranous expression in TN sample 127 (×60). |
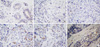 | Figure 2Geminin immunostaining. (A) Geminin low cytoplasmatic expression in normal breast cells (×60). (B) Immunonegativity for Geminin in HER2+ breast sample (×60). (C) Geminin low cytoplasmatic expression in triple-negative (TN) sample 27 (×60). (D) Geminin moderate cytoplasmatic expression in TN sample 131 (×60). (E) Geminin high cytoplasmatic expression in TN sample 7 (×60). (F) Geminin high cytoplasmatic expression in TN sample 127 (×60). |
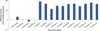 | Figure 3Geminin real time expression in breast samples. All reactions were performed in triplicate and data are expressed as the mean in the relative amount of mRNAs levels. TN=triple-negative. |
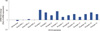 | Figure 4Prominin-1/CD133 real time expression in breast samples. All reactions were performed in triplicate and data are expressed as mean of relative amount of mRNAs levels. TN=triple-negative. |
Table 1
Clinicopathological characteristics of Triple-negative breast cancer patients and tumors and relation with CD133, Geminin, and Ki-67 expression

Table 2
Clinicopathological characteristics of non-triple-negative breast cancer patients and tumors and relation with CD133 and Geminin

Table 3
Relation between CD133 and Ki-67 markers with geminin expression in triple-negative breast cancer patients

ACKNOWLEDGEMENTS
We are grateful to Dr. Ornella Sacco, data manager of Institutional BioBank of Pascale Hospital (BBI), for expert support.
References
1. Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001. 98:10869–10874.

2. van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002. 347:1999–2009.

3. Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK, et al. Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res. 2008. 14:1368–1376.

5. Rakha EA, El-Sayed ME, Reis-Filho J, Ellis IO. Patho-biological aspects of basal-like breast cancer. Breast Cancer Res Treat. 2009. 113:411–422.

6. Thike AA, Cheok PY, Jara-Lazaro AR, Tan B, Tan P, Tan PH. Triple-negative breast cancer: clinicopathological characteristics and relationship with basal-like breast cancer. Mod Pathol. 2010. 23:123–133.

7. Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007. 26:2126–2132.

8. Honeth G, Bendahl PO, Ringnér M, Saal LH, Gruvberger-Saal SK, Lövgren K, et al. The CD44+/CD24- phenotype is enriched in basallike breast tumors. Breast Cancer Res. 2008. 10:R53.

9. Arnes JB, Collett K, Akslen LA. Independent prognostic value of the basal-like phenotype of breast cancer and associations with EGFR and candidate stem cell marker BMI-1. Histopathology. 2008. 52:370–380.

10. Chaffer CL, Weinberg RA. Cancer cell of origin: spotlight on luminal progenitors. Cell Stem Cell. 2010. 7:271–272.

11. Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009. 69:1302–1313.

12. Morel AP, Lièvre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008. 3:e2888.

13. Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005. 24:2827–2843.

14. Gonzalez MA, Tachibana KE, Chin SF, Callagy G, Madine MA, Vowler SL, et al. Geminin predicts adverse clinical outcome in breast cancer by reflecting cell-cycle progression. J Pathol. 2004. 204:121–130.

15. Dudderidge TJ, Stoeber K, Loddo M, Atkinson G, Fanshawe T, Griffiths DF, et al. Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clin Cancer Res. 2005. 11:2510–2517.

16. Dudderidge TJ, McCracken SR, Loddo M, Fanshawe TR, Kelly JD, Neal DE, et al. Mitogenic growth signalling, DNA replication licensing, and survival are linked in prostate cancer. Br J Cancer. 2007. 96:1384–1393.

17. Vargas PA, Cheng Y, Barrett AW, Craig GT, Speight PM. Expression of Mcm-2, Ki-67 and geminin in benign and malignant salivary gland tumours. J Oral Pathol Med. 2008. 37:309–318.

18. Shrestha P, Saito T, Hama S, Arifin MT, Kajiwara Y, Yamasaki F, et al. Geminin: a good prognostic factor in high-grade astrocytic brain tumors. Cancer. 2007. 109:949–956.

19. Zhu W, Depamphilis ML. Selective killing of cancer cells by suppression of geminin activity. Cancer Res. 2009. 69:4870–4877.

20. Shomori K, Nishihara K, Tamura T, Tatebe S, Horie Y, Nosaka K, et al. Geminin, Ki67, and minichromosome maintenance 2 in gastric hyperplastic polyps, adenomas, and intestinal-type carcinomas: pathobiological significance. Gastric Cancer. 2010. 13:177–185.

21. Torres-Rendon A, Roy S, Craig GT, Speight PM. Expression of Mcm2, geminin and Ki67 in normal oral mucosa, oral epithelial dysplasias and their corresponding squamous-cell carcinomas. Br J Cancer. 2009. 100:1128–1134.

22. Montanari M, Boninsegna A, Faraglia B, Coco C, Giordano A, Cittadini A, et al. Increased expression of geminin stimulates the growth of mammary epithelial cells and is a frequent event in human tumors. J Cell Physiol. 2005. 202:215–222.

23. Graeser M, McCarthy A, Lord CJ, Savage K, Hills M, Salter J, et al. A marker of homologous recombination predicts pathologic complete response to neoadjuvant chemotherapy in primary breast cancer. Clin Cancer Res. 2010. 16:6159–6168.

24. Mimeault M, Batra SK. New promising drug targets in cancer- and metastasis-initiating cells. Drug Discov Today. 2010. 15:354–364.

25. Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009. 106:13820–13825.

26. Wu Y, Wu PY. CD133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev. 2009. 18:1127–1134.
27. Cho DY, Lin SZ, Yang WK, Hsu DM, Lin HL, Lee HC, et al. The role of cancer stem cells (CD133(+)) in malignant gliomas. Cell Transplant. 2011. 20:121–125.




 PDF
PDF ePub
ePub Citation
Citation Print
Print




 XML Download
XML Download