Abstract
Purpose
Silver-enhanced in situ hybridization (SISH) is a newly developed method to evaluate HER2 gene amplification in invasive breast carcinomas. Most laboratories widely use fluorescence in situ hybridization (FISH) to evaluate the HER2 gene amplification status because FISH is a very sensitive and accurate technique. However, this technique is not the best because it requires specialized equipment and interpretation skills. We compared a new technique of SISH with FISH for assessing HER2 gene amplification in invasive breast carcinomas.
Methods
HER2 gene amplification was assessed in 165 cases of invasive breast carcinoma by FISH and SISH with constructing a tissue microarray. The tumors were assessed by the guidelines of the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP). Positivity was defined as a HER2/Chromosome 17 ratio greater than 2.2. Negativity was defined if the ratio was less than 1.8. The tumor was considered as equivocal for HER2 gene amplification if the ratio was between 1.8 and 2.2. The HER2 protein status was assessed. Immunostaining for HER2 protein was performed in a Benchmark automatic immunostaining device with using whole tissue sections.
Results
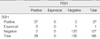
There was agreement of the HER2 gene amplification status by SISH and FISH in 162 of 165 cases, which is a concordance rate of 98.2% (κ=0.94). There were three discrepant cases, with two of them being FISH positive and SISH negative (one case was IHC negative and one case was IHC positive) and one case was FISH negative and SISH equivocal.
The HER2/neu oncogene is a member of the epidermal growth factor receptor family, and its amplification is known to be one of the most common genetic alterations associated with human breast cancer.(1) The detection of HER2 gene amplification is necessary for the proper selection of breast cancer patients responsive to the humanized anti-HER2 monoclonal antibody Trastuzumab (Herceptin® Genentech, South San Francisco, USA)(2,3) and the small molecule dual HER1/HER2 tyrosine kinase inhibitor Lapatinib (Tykerb®, GlaxoSmithKline, Philadelphia, USA). (4,5)
Immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH) are used to determine HER2 gene amplification. In 2007, the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) made a guideline of recommendations for HER2 testing in breast cancer.(6) They suggest an algorithm defining positive, equivocal, and negative values for both HER2 protein expression by IHC and gene amplification by FISH and a new silver-enhanced in situ hybridization (SISH). They emphasized that the performance of a testing algorithm should rely on an accurate, reproducible assay. They specified elements to reduce assay variation such as specimen handling, assay exclusion, and reporting criteria. FISH is a popular technique to determine the status of HER2 gene amplification on a genomic level. However, FISH has disadvantages which include a longer staining and scoring time, training of personnel for the interpretation of slides, use of fluorescence microscopy, and impermanence of the fluorochrome stain.(7) A newly developed SISH is a fully automated assay providing permanent stained slides in six hours that can be interpreted by conventional light microscopy.(8) The aim of this study was to compare HER2 gene amplification by both methods in a series of invasive breast carcinomas.
Two hundred and one consecutive breast cancer cases which were diagnosed and treated surgically during 2003 and 2004 at the Asan Medical Center, Seoul were selected for this study. In all cases, samples were formalin-fixed, paraffin-embedded and processed in a pathology laboratory according to the institutional standardized protocols.
Immunostaining of HER2 was performed in a Benchmark automatic immunostaining device (Ventana Medical System, Tucson, USA) using formalin-fixed, paraffin-embedded tissue sections. Five µm thick sections were obtained with a microtome, transferred into adhesive slides, and dried at 62.8℃ for 30 min. After incubation with primary antibody against HER2 (1:500 dilution, DAKO, Glostrup, Denmark), immunodetection was performed with biotinylated antimouse immunoglobulin followed by peroxidase-labeled streptavidin using a labeled streptavidin biotin kit with 3,3'-diaminobenzidine chromogen as the substrate. The primary antibody incubation step was omitted in the negative control. The normal breast tissues entrapped within the block and appropriate control tissues were used as positive controls. Slides were counterstained with Harris hematoxylin.
Formalin-fixed, paraffin-embedded tissue samples were arrayed using a tissue-arraying instrument. Briefly, representative areas of each tumor were selected and marked on the H&E slide, and its corresponding tissue block was sampled. The designated zone of each donor block was punched with a tissue cylinder 1 mm in diameter, and the sample was transferred to a recipient block. Each sample was arrayed in duplicate to minimize tissue loss and overcome tumor heterogeneity. The slides were prepared from archived paraffin blocks and were processed in parallel for FISH (PathVysion® HER2 DNA Probe Kit, Abbott/Vysis, Des Plaines, USA), SISH (INFORM® HER2 DNA probe and ultraView™ SISH Detection Kit, Ventana Medical Systems, Tucson, USA).
For SISH, 5 µm-thick sections from the microarray block were prepared. The automated SISH of slides were performed according to the manufacturer's protocols for INFORM HER2 DNA and chromosome 17 probes.(9) Both probes were labeled with dinitrophenol (DNP) and optimally formulated for use with the ultraView SISH Detection Kit and accessory reagents from Ventana's Benchmark® series of automated slide stainers. The HER2 DNA probe was denatured at 95℃ for 12 min and hybridization was performed at 52℃ for 2 hr. After hybridization, appropriate stringency washes (3 times at 72℃) were performed. The chromosome 17 probe was denatured at 95℃ for 12 min and hybridization was performed at 44℃ for 2 hr. After hybridization, appropriate stringency washes (3 times at 59℃) were performed. The HER2 and chromosome 17 DNP-labeled probes were visualized using the rabbit anti-DNP primary antibody and the ultraView SISH Detection Kit which contains a goat anti-rabbit antibody conjugated to horseradish peroxidase utilized as the chromogenic enzyme. Silver precipitation is deposited in the nuclei and a single copy of the HER2 gene is visualized as a black dot. The specimen is then counterstained with Harris hematoxylin.
Consecutive sections from the microarray blocks were cut at 5 µm thickness and mounted on SuperFrost +/+ slides. Deparaffinizing, pre-treatment and protease digestion procedures followed the Abbott PathVysion HER2 DNA Probe Kit protocol (Abbott Laboratories, Abbott Park, USA) with additional monitoring for the progress of proteolytic digestion by propidium iodide staining. Probe mixes were hybridized at 37℃ between 14 and 18 hr. After hybridizations, slides were washed in 2X SSC/0.3% NP-40 at 72℃ for 30 min, air dried and counterstained with DAPI.
SISH signals were visualized as single copies, multiple copies, and clusters. A discrete dot was counted as a single copy of HER2 or Chr17. The size of these single dots was used as a reference to determine the relative number of amplified copies in the cancer nuclei. In some nuclei, clusters of dots representing many copies of HER2 gene were apparent. A small cluster of multiple signals was counted as six signals and a large cluster as 12 signals. Enumeration of HER2 and Chr17 signals in 20 nuclei was done within a target area and the HER2/Chr17 ratio was calculated. Cases with a HER2/Chr17 ratio less than 1.8 were negative for HER2 gene amplification while cases with a HER2/Chr17 ratio greater than 2.2 were positive for HER2 gene amplification. If a HER2/Chr17 ratio was either equal to or fell between 1.8 and 2.2, we counted the number of signals in another 20 additional nuclei in a second target area. The HER2/Chr17 ratio was then calculated from both target areas (40 cells).
SISH scoring critera; 1) Negative for HER2 gene amplification: HER2/Chr17 ratio less than 1.8. 2) Equivocal for HER2 gene amplification: HER2/Chr17 ratio equal to or between 1.8 and 2.2. 3) Positive for HER2 gene amplification: HER2/Chr17 ratio greater than 2.2.
Typical examples of a non-amplified case and amplified case are given in Figure 1.
FISH scoring criteria; 1) Negative for HER2 gene amplification: less than 4.0 HER2 gene copies per nucleus or a HER2/Chr17 ratio less than 1.8. 2) Equivocal for HER2 gene amplification: HER2/Chr17 ratio between 1.8 and 2.2. 3) Positive for HER2 gene amplification: more than 6 HER2 gene copies per nucleus or as a HER2/Chr17 ratio greater than 2.2.
IHC scoring criteria; 1) Negative for HER2 protein: no staining or weak, incomplete membrane staining in any proportion of the tumor cells. 2) Equivocal for HER2 protein: complete membrane staining that is either non-uniform or weak in intensity but with obvious circumferential distribution in at least 10% of cells. 3) Positive for HER2 protein: uniform intense membrane staining of >30% of the invasive tumor cells.
From 201 cases, 165 were informative for both SISH and FISH (Table 1). Twenty five cases were excluded because of the absence of tumor cells in the microarray tissue and 11 cases for poor staining by SISH. The status of HER2 gene amplification status by SISH and FISH was in agreement in 162 out of 165 cases representing a concordance of 98.2% (κ=0.94) (Table 1). There were three discrepant cases, two of them representing FISH positive, SISH negative (one case is IHC negative and one case is IHC positive) and one representing FISH negative, SISH equivocal.
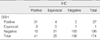
The status of HER2 gene amplification except for equivocal cases by SISH and HER2 protein expression by IHC was in agreement in 136 out of 148 cases representing a concordance of 91.9% (κ=0.78) (Table 2). The status of HER2 gene amplification except for equivocal cases by FISH and HER2 protein expression by IHC was in agreement in 147 out of 159 cases representing a concordance of 92.5% (κ=0.81) (Table 3).
The major aim of this study was to compare SISH to FISH assays presently used in diagnostic pathology laboratories and thus validate its use as a routine diagnostic method for assessing HER2 status in breast cancers. Recently, a few manuscripts have been published about the excellent reproducibility and efficacy of HER2 SISH technique and interobserver interpretation. Carbone et al.(10) reported excellent reproducibility and efficacy of HER2 SISH staining and interobserver interpretation (Kw=0.91) among five different institutions and high concordance between consensus IHC and consensus SISH (96.6%), FISH (97.8%), and chromogenic ISH (96.6%). Dietel et al.(9) reported high concordance between FISH and SISH (96.0%, κ=0.754 ) and a low interobserver variability in the interpretation of SISH.
The current study also showed that SISH yielded a high concordance with FISH (98.2%) and IHC (91.9%). The 98.2% concordance between FISH and SISH meets the ASCO/CAP requirements for test validation of >95% concordance for amplified versus non-amplified cases. The ASCO/CAP guidelines recommended that laboratories should show 95% concordance with another validated test for positive and negative assay values to perform HER2 testing. The panel strongly recommends validation of laboratory assay or modifications, use of standardized operating procedures, and compliance with new testing criteria to be monitored with the use of stringent laboratory accreditation standards, proficiency testing, and competency assessment.(6)
FISH has long been held to be a gold standard technique for assessing HER2 status on a genomic level. It has high sensitivity (96.5%) and specificity (100%) for detecting HER2 gene amplification.(11) FISH has several advantages such as it can be done with only a small volume of tumor samples and on formalin-fixed and paraffin-embedded tissue samples with tissue preparation having little or no effect on the testing. It also permits direct visualization of gene amplification in the nuclei and provides an objective count of the genes and chromosomes on a cell-by-cell basis. However, there are several disadvantages of FISH. It requires a fluorescence microscope and special training for interpretation. It also may be difficult to visualize the morphologic features of the tumor cells and to separate in situ from invasive carcinoma. In addition, fluorescence fades quickly and, therefore, the FISH slides are not permanent and as a result the data is lost. Besides, FISH is a time-consuming (2-3 days), laborious, non-automated and expensive assay. Further, use of FISH exclusively as the primary method for determining the status of HER2 gene amplification may also be problematic.
As alternative assays that can overcome the disadvantages of FISH, brightfield ISH including chromogenic in situ hybridization (CISH) and SISH have been developed.(6) CISH has been validated as an acceptable assay allowing for the detection of HER2 gene copies using a simple immunohistochemistry-like peroxidase reaction, enumeration of gene copy number with simultaneous histologic examination by regular brightfield microscopy and permanent storage since CISH signal intensity does not diminish over time.(12,13) But it requires manual processing and overnight hybridization. SISH has not only the benefits of using regular brightfield microscopy and permanent signal intensity but also a six hour automated protocol saving more time and requiring less effort than CISH.
In this study, 11 out of 186 cases having evaluable tumor cells on the slides of SISH (5.9%) showed poor SISH staining. The tumor cell and internal control cells (fibroblast and endothelial cells) had no signals. Also in same tissue microarray slide, there was some heterogeneous staining intensity in the sample. Judging from the fact that each two cores from the same case had similar intensity, the poor staining and heterogeneous intensity might be caused by a problem in the procedure for tissue processing.
The response of the Herceptin in the discrepant cases may be important to determine the reliability of the test. However, because this study was done with the patients who were treated before Herceptin was used for the breast cancer treatment, the response could not be followed.
Figures and Tables
 | Figure 1Typical example of a non-amplified case and amplified case as demonstrated by Silver-enhanced in situ hybridization (SISH). (A) Two single dots represent each Chr17, (B) non-amplified HER2 case showed 2 HER2 signals in tumor cells (arrow), (C) amplified HER2 case showed clusters of HER2 signals in tumor cells (arrow). Note the fibroblasts (arrow head) as internal control (×1,000). |
References
1. Sahin AA. Biologic and clinical significance of HER2/neu (cerbB-2) in breast cancer. Adv Anat Pathol. 2000. 7:158–166.

2. Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999. 17:2639–2648.

3. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001. 344:783–792.

4. Cameron D, Casey M, Press M, Lindquist D, Pienkowski T, Romieu CG, et al. A phase III randomized comparison of lapatinib plus capecitabine versus capecitabine alone in women with advanced breast cancer that has progressed on trastuzumab: updated efficacy and biomarker analyses. Breast Cancer Res Treat. 2008. 112:533–543.

5. Geyer CE, Forster J, Lindquist D, Chan S, Romieu CG, Pienkowski T, et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N Engl J Med. 2006. 355:2733–2743.

6. Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007. 25:118–145.

7. Dowsett M, Hanna WM, Kockx M, Penault-Llorca F, Rüschoff J, Gutjahr T, et al. Standardization of HER2 testing: results of an international proficiency-testing ring study. Mod Pathol. 2007. 20:584–591.

8. Cell Markers and Cytogenetics Committees College Of American Pathologists. Clinical laboratory assays for HER2/neu amplification and overexpression: quality assurance, standardization, and proficiency testing. Arch Pathol Lab Med. 2002. 126:803–808.
9. Dietel M, Ellis IO, Höfler H, Kreipe H, Moch H, Dankof A, et al. Comparison of automated silver enhanced in situ hybridisation (SISH) and fluorescence ISH (FISH) for the validation of HER2 gene status in breast carcinoma according to the guidelines of the American Society of Clinical Oncology and the College of American Pathologists. Virchows Arch. 2007. 451:19–25.

10. Carbone A, Botti G, Gloghini A, Simone G, Truini M, Curcio MP, et al. Delineation of HER2 gene status in breast carcinoma by silver in situ hybridization is reproducible among laboratories and pathologists. J Mol Diagn. 2008. 10:527–536.

11. Pauletti G, Godolphin W, Press MF, Slamon DJ. Detection and quantitation of HER2/neu gene amplification in human breast cancer archival material using fluorescence in situ hybridization. Oncogene. 1996. 13:63–72.




 PDF
PDF ePub
ePub Citation
Citation Print
Print



 XML Download
XML Download