INTRODUCTION
Breast carcinoma is one of the most common malignancies affecting women worldwide. Recent research for tumor biology has resulted in new insights in the regulation of cell kinetics, invasion and metastasis.(1-3) But, the pathogenic mechanism underlying development and progression of breast carcinoma is still largely unknown. The most important prognostic factor is the stage of tumor at diagnosis as classified in TNM tumor staging, including the involvement of axillary regional lymph nodes and metastases of distant organs.(4, 5) Until now, we have used a few immunohistochemically detectable markers relevant for the pathologic process and the prognosis of breast cancer, although it has not been satisfactory. Therefore, it is important to identify a potential tumor marker that is associated with pathophysiologic processes of breast carcinoma.
The genes N-Myc downstream regulated gene 1-4 (NDRG1-4), which comprises the family of proteins homologous to N-Myc downstream-regulated protein 1, have been implicated in stress responses, putative roles in neural differentiation, synapse formation, and axon survival. NDRG1, also called Drg1, Cap43, rit42, TDD5, and Ndr1, which is the most widely expressed and best characterized member, is mutated in hereditary motor and sensory neuropathy-Lom, a form of Charcot-Marie Tooth disease.(6) The expression of NDRG1 is inhibited by N-Myc,(7) and the protein is repressed in transformed cells and up-regulated in growth-arrested differentiating cells.(8) Although NDRG2 is in fact N-Myc-independent,(9) it appears to inhibit cell proliferation and promote differentiation.(10, 11) NDRG2 is highly expressed in the adult brain, heart, salivary gland and skeletal muscle but is undetectable in proliferative tissues such as the colon and bone marrow. It is expressed during the differentiation of human monocyte-derived dendritic cells and its gene expression is differentially regulated by maturation-inducing stimuli.(11) NDRG 3 and NDRG 4 expression are restricted to the brain and heart,(12) suggesting that it might display different specific functions in distinct tissues. However, the molecular basis of the tumor-suppressor activity of human NDRG2 (hNDRG2) is unknown in a series of human breast carcinoma at the present time. Herein, the authors tried to reveal the correlations between the expression of hNDRG2 and the various clinicopathologic prognostic factors such as age, lymph node metastases, estrogen receptor (ER), and progesterone receptor (PR), in invasive ductal carcinoma of the breast.
To investigate the functional and clinicopathologic roles of the hNDRG2 protein in tumorigenesis of human breast carcinoma, the authors used eukaryotic transfection to manipulate the expression of hNDRG2 in MDA-MB-231 human breast cancer cell line and examined an intracellular biological role in regulation of hNDRG2 overexpression, together with the possible correlation between the expression of hNDRG2 protein in association with the various clinocopathologic prognostic parameters in 67 human breast carcinomas.
METHODS
1. Cell culture and Western blot analysis
The cells used in our studies were the human breast cancer cell lines; MDA-MB-231, MCF-7 and colon adenocarcinoma cell lines; HCT116, Colo205, HT29, SW620 and KM12 (ATCC, Rockville, USA). The cells were cultured in appropriate media, as follows: RPMI 1640 medium (Sigma Chemical Co., St. Louis, USA); each was supplemented with 10% fetal bovine serum (Life Technologies Inc., Grand Island, USA) and 1% antibiotic-antimycotic solution (Life Technologies Inc., Grand Island, USA). The cells were kept at 37℃ in a humidified incubator which was maintained with 5% CO2.
2. Patients and specimens
The breast carcinoma samples were obtained from patients who underwent routine surgery for breast cancer at the Department of Surgery, Chonbuk National University Hospital in 2002-2004. Patients included in the study had an axillary dissection that sampled at least five lymph nodes. The cancerous breast and paired normal breast tissues taken from a site distant from the tumorous lesion were snap frozen and stored in liquid nitrogen till further use. For the immunohistochemical study, some of these tissue specimens were fixed in 10% neutralized buffered formalin solution for 24 hr. Each patient's clinical status was classified according to the pathological grade of the tumor size, lymph node, and metastasis (pTNM) classification system.(13)
3. Reverse transcription-polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted from 10 breast carcinoma tissues and non-tumorous tissues of the patients using the AGPC method.(14) Reverse transcription reactions were done with a cDNA synthesis kit (Stratagene, La Jolla, USA) following the instruction manual. The cDNA was synthesized with 7 µg of total RNA and oligo (dT) primer in 50 µL of a solution containing reverse transcriptase. The reverse-transcribed samples were used as templates for amplification of hNDRG2 and G3APDH gene, which was used as an internal quantitative control. For the PCR reactions, the following primers were used: sense primer of hNDRG2 5'-CGGGATCCATGGCGTGCCCTCTGGAG-3' and antisense primer 5'-CCGCTCGAGTCATTTCTTCCTGGGCTG-3'. With respect to G3APDH, the primer sequences were as follows: sense primer 5'-CCCCTGGCCAAGGTCATCCATGACAACTTT-3' and antisense primer 5'-GGCCATGAGGTCCACCACCCTGTTGCTGTA-3'. The PCR reactions were performed following the cycling parameters on a Minicycler™ PCR system (MJ Research Inc., Watertown, USA): 10 min at 94℃ followed by 25 cycles of 1 min at 94℃, 1 min at 55℃, 1 min at 72℃, and a final cycle at 72℃ for 10 min. Quantitation of the PCR products was scanned and performed using a Quantity One program (Bio-Rad, Hercules, USA). Increased expression of the hNDRG2 mRNA in tumor tissue was defined as more than about 2-3 times higher than that seen in normal breast tissue.
4. Western blot analysis
Protein isolation and Western blotting analysis were performed as described.(15) Immunostaining and protein band visualization with the ECL system was carried out according to the manufacturer's protocol. MDA-MB-231 breast cancer cells were grown to 60-70% confluence and rinsed 2 times with phosphate-buffered saline (PBS). And then lysed in RIPA buffer (1X PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with a protease inhibitor cocktail added. Total proteins were normalized, NuPage sample buffer (Invitrogen, Carlsbad, USA) was added and 30 µg of protein were electrophoresed on 4-12% Bis/Tris NuPage gradient gels (Invitrogen, Carlsbad, USA). Proteins were transferred onto nitrocellulose membrane (Invitrogen, Carlsbad, USA). The membranes were blocked overnight in 10% Tris-buffered saline/0.05% Tween-20 (TBS-T) at 4℃ and then probed with the appropriate antibody in 2% skim milk in TBS-T. The antibodies used were hNDRG2 and beta-actin. After washing, the blots were treated with the appropriate secondary horseradish peroxidase-conjugated antibody and washed. Proteins were detected using enhanced chemiluminescence system (Amersham, Arlington Heights, USA) and exposed to Hyperfilm-MP (Amersham, Arlington Heights, USA).
5. Immunohistochemical analysis
Immunohistochemistry was performed to study altered protein expression in 67 human breast carcinoma tissues. Mouse monoclonal antibody against hNDRG2 was used as primary antibody. Also, commercially available mouse monoclonal antibodies against p53 DO-7 (Dako, Glostrup, Denmark, dilution: 1:100), c-erbB2 (Dako, Kyoto, Japan, dilution: 1:100) Estrogen Receptor (Dako, Kyoto, Japan, dilution: 1:50), and Progesterone Receptor (Dako, Kyoto, Japan, dilution: 1:10) were used as primary antibodies. A paraffin section of the breast carcinoma tissue was deparaffinized and rehydrated in graded alcohol. Antigenic enhancement was performed by submerging in 1x citrate buffer (pH 6.0) and microwaving. The sections were then treated with 3% hydrogen peroxide in methanol to quench the endogenous peroxidase activity, followed by incubation with 1% BSA to block the non-specific binding, respectively. The primary polyclonal anti-Bmi-1 antibody was incubated for 90 min at room temperature. After washing, the tissue section was then reacted with the biotinylated anti-goat secondary antibody, followed by incubation with streptavidin-horseradish-peroxidase complex. The tissue section was immersed in 3-amino-9-ethyl carbazole as a substrate. In negative controls, non-immune goat IgG of the same isotype or antibody dilution solution replaced the primary antibody.
Each section was evaluated by at least two independent observers, and moderate to strong cell membrane or cytoplasmic staining was considered as a positive reaction. The distribution of hNDRG2 was scored on a semiquantitative scale, as follows: negative (<10% of tumor positive), focally positive (10-50% of tumor positive), and diffusely positive (>50% of tumor positive). For Estrogen Receptor, a nuclear staining in >25% of the cells was considered Estrogen Receptor positive, and similarly for Progesterone Receptor. A nuclear staining in >10% of the cells were considered positive for p53.
6. Transfection and selection of hNDRG2 over-expressing stable cells
MDA-MB-231 breast cancer cells were transfected with appropriate plasmids using the Fugene 6 reagent (Boehringer Mannheim, Mannheim, Germany) as recommended by the manufacturer. Expression plasmids encoding the nucleotide sequence corresponding to the full open reading frame (ORF) for hNDRG2 were subcloned as 5' BamHI/3' XhoI fragment into pcDNA3.1 (+) (Invitrogen, Carlsbad, USA) for transfection. MDA-MB-231 cells were plated at 1×106 cells/well in a 10 cm plate. One days later, the cells were transfected with wild type hNDRG2 (5 µg) or pcDNA3.1 (+) empty vector using the Fugene 6 reagent (Boehringer Mannheim, Mannheim, Germany). For the selection of transfected cells, cells were treated with G418 after transfection for 48 hr. The selected cells were collected and confirmed by western blot analysis.
7. Growth study
MDA-MB-231 breast cancer cells which transfected hNDRG2 and Mock control cells were seed at a density of 2×105 per well in 6-well plates and cultured for five days. Cells were then trypsinized, stained with 0.4% trypan blue (Invitrogen, Carlsbad, USA), and counted daily using a hemocytometer (Fisher Scientific, Allentown, USA). Only unstained, live cells were counted.
8. Preparation of cDNA probe and microarray hybridization
The oligo microarray containing a set of 22,746 human oligo (Illumina, Inc., San Diego, USA) was provided by GenomicTree, Inc (Daejon, Korea). The synthesis of target cDNA probes and hybridization were performed according to previously described.(16) Each 5 µg total RNA was reverse transcribed in the presence of Cy3 or Cy5-dUTP (NEN Life Sciences, Boston, USA) at 42℃ for 2 hr. Control RNA was labeled with fluorescent Cy3-dUTP and Test condition RNA was labeled with fluorescent Cy5-dUTP. Both the Cy3 and Cy5-labeled cDNA were purified using PCR purification kit (Qiagen, Valencia, USA) as recommended by manufacturer. The purified cDNA was resuspended in 100 µL of hybridization solution containing 5X SSC, 0.1% SDS, 30% formamide, 20 µg of Human Cot-1 DNA, 20 µg of poly A RNA and 20 µg of Yeast tRNA (Invitrogen, Carlsbad, USA). The hybridization mixtures were heated at 100℃ for 2-3 min and directly pipetted onto microarrays. The arrays hybridized at 42℃ for 12-16 hr in the humidified hybridization chamber (GenomicTree Inc., Daejon, Korea). The hybridized miroarrays were washed with 2×SSC/0.1% SDS for 5 min, 0.1X SSC/0.1% SDS for 10 min, and 0.1×SSC for 2 min two times. The washed microarrays were immediately dried using the microarray centrifuge (GenomicTree Inc., Korea). After hybridization, microarray slides were imaged using laser scanner (Axon 4000B, Axon Instruments Inc., Foster, USA). The signal and background fluorescence intensities were calculated for each probe spot by averaging the intensities of every pixel inside the target region using GenePix Pro 4.0 software (Axon Instruments Inc., Foster, USA). Spots were excluded from analysis due to obvious abnormalities. All data normalization, statistical analysis and cluster analysis were performed using GeneSpring 7.2 (Agilent, Palo Alto, USA). The fold change was calculated by dividing the median of normalized red channel intensity by the median of normalized green channel intensity.
9. DNA methylation analysis of the hNDRG2 gene
For the evaluation of DNA methylation state, the hNDRG2 promoter was analyzed for CpG site methylation using sodium bisulfite modified genomic DNA. Sodium bisulfite treatment of genomic DNA was carried out as described by Herman et al.(14)
In brief, 1 µg DNA was denatured with 0.2 M NaOH in a total volume of 50 µL. After the addition of 350 µL of 3.6 M sodium bisulfite containing 1 mM hydroquinone at pH 5, the samples were incubated for 16 hr at 55℃ in dark. The modified DNA was recovered with 5 µL glassmilk (BIO 101, Irvine, USA) and 800 µL of 6 M NaCl. The glassmilk catching modified DNA was washed three tines with 70% ethanol at room temperature, treated with 0.3 M NaOH/90% ethanol, and washed twice with 90% ethanol. The DNA was finally eluted from the dried pellet with 30 µL 1 mM Tris-HCl (pH 8.0) for 15 min at 55℃. The bisulfite-modified DNA (5 µL) were subjected to methylation specific PCR (MSP) using two sets of primer specific for methylated and unmethylated DNA. Two overlapping fragments from the hNDRG2 promoter region were PCR-amplified from sodium bisulfite-modified DNA using the following primers, as described by Herman et al.(17): P1-sense, 5'-TTTTCGAGGGGTATAAGGAGAGTTTATTTT-3' and P1-antisense, 5'-CCAAAAACTCTAACTCCTAAATAAACA-3' (320 bp product); P2-sense, 5'-TTAAGGATATTGCGTTTTTTTTAAGTTTTTATTTT-3' and P2-antisense, 5'-AAAATTCCGACTCCCTCGTACCCAAAA-3'(335 bp product). PCR amplification was carried out for 43 cycles and controls without DNA were performed for each set of PCRs. The PCR production (10 µL) was directly loaded onto 2% agarose gels containing ethidium bromide, and directly visualized under UV illumination.
10. Statistical analysis
The relationship between the results of the immunohistochemical study and the clinicopathologic parameters was performed using the SAS® software package (version 8.01; SAS Institute, Cary, USA). Univariate and multivariate analyses were carried out using the proc logistic module. In all cases, the exact mid-p adjusted p-values were reported and a p-value <0.05 was considered to be statistically significant.
RESULTS
1. hNDRG2 expression in human invasive ductal breast cancer
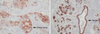
The expressed patterns of the hNDRG2 in human normal breast tissue and carcinomatous tissue were examined through immunohistochemical staining. Immunohistochemical examination of the tumor tissues using monoclonal antibody specific to hNDRG2 showed that 15 of 67 (22%) breast tumorous tissues samples expressed hNDRG2 gene, and that hNDRG2 was largely distributed in the cell membrane, but some cytoplasmic areas of tumor cells (Fig 1A). Normal breast tissue including the ductal and lobular epithelial cells and ductal carcinoma in situ tissue from patients showed strong hNDRG2 expression in cytoplasm and plasma membrane of the cells (Fig 1B). Interestingly, hNDRG2 protein was significantly decreased in 52 cases (78%) of 67 breast carcinomas, compared with that in normal mucosal tissue from cancer patients (Fig 2). The expression patterns of hNDRG2 proteins in 67 human breast tumors examined as negative/focal, moderately and diffuse positive immunoreactivities in each case, respectively.
The relative levels of expression of the NDRG2 mRNA in the human breast carcinoma tissues were compared with those of non-tumorous tissues. The author examined the level of hNDRG2 gene products using RT-PCR analysis in 10 matched breast cancer and adjacent normal breast samples. The results showed that a significant difference at mRNA levels was not detected between cancerous tissue and normal breast tissue, although mRNA level of hNDRG2 were increased in some breast cancers (data not shown).
2. Establishment of stable hNDRG2 over-expressing cell line and hNDRG2 over-expression leads to a decrease in cell proliferation
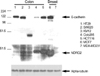
To elucidate the role of hNDRG2 in human breast carcinoma, we performed a eukaryotic transfection to manipulate the expression of hNDRG2 in MDA-MB-231 breast cancer cell line. Prior to eukaryotic transfection, we performed western blot analysis for the evaluation of hNDRG2 expression level in two breast tumor cell lines and five colon tumor cell lines (Fig 3). Of these tumor cell lines, MDA-MB-231 cells and SW620 cells showed low basal levels of hNDRG2 protein and MDA-MB-231 cells was selected for eukaryotic transfection. Ectopic overexpression of hNDRG2 showed a decrease in cell proliferation. But, the other pathologic findings such as a change in the morphologic phenotype were not feasible. On continuous cell culture, we found that hNDRG2-transfected cells showed decrease in cell number compared with Mock control (Fig 4). Under the serum-dependent manner and cell density culture conditions, hNDRG2 protein was expressed as two different molecular weights in western blot analysis. Especially, hNDRG2 protein was phosphorylated in a 48-kDa sized band (Fig 5).
3. Oligo microarray data acquisition and analysis
We investigated the status of differentially-expressed genes between Mock control and hNDRG2 over-expressing MDA-MB-231 cells. Of the 24,000 genes, 320 genes showed a net difference by more than 4-fold in expression intensity between Mock control-array and hNDRG2-array (Fig 6). Genes whose expression was altered by more than 4-fold are listed in Table 1, 2. Sixty four genes showed increase in expression while 256 genes were repressed by hNDRG2 overexpression. The genes whose expression was altered by hNDRG2 overexpression may be grouped into four categories: cell surface protein, signaling protein, nuclear protein, and others.
4. DNA methylation analysis of the hNDRG2 gene
The author examined the promoter hypermethylation of the hNDRG2 gene using two sets of primers specific for methylation specific PCR on bisulfite modified DNA. In human tumor cell lines, MDA-MB-231 breast cancer cell line and SW620 colon cancer cell line showed both methylated and unmethylated DNA PCR products (Fig 7A). In corresponding normal breast tissues, unmethylated DNA of the NDRG2 gene was amplified in all 11 samples but methylated DNA was not amplified in any cases. Also, hypermethylation was not found in all 11 tumorous samples examined, although unmethylated DNA of the NDRG2 gene was amplified in all samples (Fig 7B).
5. Relationship between hNDRG2 expression and clinicopathological parameters in invasive ductal breast cancer
Relationship of hNDRG2 expression between human normal breast tissue and invasive breast cancer tissue was analyzed (Table 3). hNDRG2 was significantly down-regulated in the invasive ductal carcinomatous tissue, compared to the normal ductal and lobular tissues (p<0.0001). The author investigated the hNDRG2 expression and various clinicopathological parameters to elucidate the roles of the NDRG2 gene in tumorigenesis of the breast cancer. As the data summarized in Table 4, our study showed that there was a correlation between the hNDRG2 positivity and tumor size, and histologic grade. But clinicopathological parameters such as axillary lymph node metastases, age, and p53 positive status were not correlated in univariate and multivariate analysis. These findings suggested the possibility that hNDRG2 protein might play a role in the development and progression of human invasive ductal breast cancer.
DISCUSSION
Human breast cancer has been increasing steadily and many patients with this malignancy have suffered during the last several decades. Available clinicopathologic prognostic indicators are not accurate; despite axillary nodal status being the most important factor that determines the overall survival in patients with breast cancer, approximately 25-30% of node-negative cases eventually relapse.(5) Thus, it is important to identify a biological genetic molecular marker that is associated with pathophysiologic processes of human breast cancer.
The author have identified a significant decreased novel protein, designated hNDRG2, by using an immunohistochemical technique in human solid tumor. The results presented here are significant in two respects. First, the down-regulation of hNDRG2 expression is a common histopathologic feature in the human solid tumor studied, especially invasive breast carcinoma. Second, the over-expression of hNDRG2 protein in MDA-MB-231 breast carcinoma cell line induced a decrease of cell proliferation. These findings strongly imply that hNDRG2 protein has functions closely associated with the tumor development and progression in human breast cancer.
The NDRG2 protein belongs to a family of highly conserved and four closely related proteins, whose intracellular functions are currently unknown.(18, 19) Two isoforms of NDRG2 have been described.(20) The insertion of 42 base pairs into the mRNA by alternative splicing resulted in a protein containing 14 additional amino acids, which was described as NDRG2ins. Structurally, it has been noted to harbor a central alpha/beta-hydrolase fold domain.(12, 18) However, the catalytic triad residues in the NDRG family members do not conform to the consensus for alpha/beta-hydrolase. The genes NDRG1-4, which comprises the family of proteins homologous to N-Myc downstream-regulated protein 1, have been implicated in stress responses, putative roles in neural differentiation, synapse formation, and axon survival. hNDRG1, also called Drg1, Cap43, rit42, TDD5, and Ndr1, which is the most widely expressed and best characterized member, is mutated in hereditary motor and sensory neuropathy-Lom, a form of Charcot-Marie Tooth disease.(6) The expression of hNDRG1 is inhibited by N-Myc,(7) and the protein is repressed in transformed cells and up-regulated in growth-arrested differentiating cells.(8) Although hNDRG2 is in fact N-Myc-independent,(9) it appears to inhibit cell proliferation.(10) Recently, it is also expressed during the differentiation of human monocyte-derived dendritic cells and its gene expression is differentially regulated by maturation-inducing stimuli.(11) NDRG2 is highly expressed in the adult brain, heart, salivary gland and skeletal muscle but is undetectable in proliferative tissues such as the colon and bone marrow. NDRG3 and NDRG4 expressions are restricted to the brain and heart,(12) suggesting that it might display different specific functions in distinct tissues. But, the molecular basis of the tumor-suppressor and differentiation activity of hNDRG2 is unknown in the human epithelial tumor at the present time.
NDRG2 gene has been reported as a gene specifically expressed in murine and human cells. Expression of hNDRG2 protein in human cancer was reported in only some papers and little is known about the intracellular role of hNDRG2 protein in the pathophysiologic process of tumorigenesis in human breast cancer. Human breast adenocarcinoma is one of the major causes of morbidity and mortality worldwide, its tumorigenic mechanism is a multi-step process related to the genetic instability associated with genetic alterations. Currently, the most important prognostic factor is the stage of tumor at diagnosis as classified in pTNM tumor staging, including involvement of regional lymph nodes and infiltration of distant internal organs. Until now, we have used a few immunohistochemically detectable markers relevant for the pathologic process and the prognosis of breast cancer, although it has not been satisfactory. Therefore, it is important to identify a potential tumor marker that is associated with pathophysiologic processes of colorectal carcinoma. In the present study, hNDRG2 was ubiquitously expressed and found in specific cell types such as epithelial and dendritic cells in human breast tumorous specimen. The author examined the level of hNDRG2 gene products using RT-PCR analysis in 10 matched breast cancerous and adjacent normal breast samples. The results showed that a significant difference at mRNA levels was not detected between cancerous tissue and normal mucosal tissue, although mRNA level of hNDRG2 were increased in some colorectal cancers (data not shown). I think that this result might be caused by the superficial sampling in the breast cancer specimen. On immunohistochemical staining, the proportion of hNDRG2 over-expression was only 32% at protein levels. It was largely detected in cytoplasm of the normal mucosal epithelial cells but detected in cytoplasm and cell membrane in the tumor cells of breast cancer. In addition, the author analyzed the possible correlation for the expression of hNDRG2 protein in association with the various clinocopathologic prognostic parameters in 67 human breast adenocarcinomas. As a result, the increased level of expression of hNDRG2 protein significantly correlated with tumor differentiation but inversely correlated with TNM stage in univariate and multivariate analyses, respectively. hNDRG2 expression was not correlated with the other clinicopathologic parameters tested in the human breast cancer. Therefore, hNDRG2 protein indicates a biologic tumor suppressor and one of the useful prognostic tumor markers for human breast cancer.
To investigate the functional and patholophysiologic roles of the hNDRG2 protein in tumorigenesis of human breast carcinoma, the author used eukaryotic transfection system to manipulate the expression of hNDRG2 in MDA-MB-231 human breast cancer cell line, which showed the basal level of endogenous hNDRG2, and examined an intracellular biological role in regulation of hNDRG2 expression. Ectopic overexpression of the hNDRG2 showed a decrease of cell proliferation compared with mock-MDA-MB-231 control. However, other pathologic findings such as morphologic phenotypic change were not found. In addition, it was confirmed that cyclin D level was decreased in oligo-microarray analysis, suggesting the possibility of its transcriptional down-regulation. Eukaryotic cell cycle progression is promoted by the activity of phase-specific kinase complexes composed of cyclins and cyclin-dependent kinases.
hNDRG2 has been identified as a new component of the insulin signaling cascade in skeletal muscle, most likely directly phosphorylated by Akt.(21) The protein is also a specific substrate for protein kinase C (PKC) and a site of cross-talk between the two signaling pathways, and hence of relevance to the development of PKC-mediated insulin resistance in this tissue. The protein kinase Akt mediates several metabolic and mitogenic effects of insulin, whereas activation of PKC isoforms has been implicated in the inhibition of insulin action.(22-24) hNDRG2 expressed two different molecular weights in western blot analysis. Especially, hNDRG2 was phosphorylated in a 48-kDa sized protein. hNDRG2 protein was changed at its molecular size level under the several culture conditions, such as serum-dependent manner or cell density. Indeed, it is most likely that phosphorylation of hNDRG2 may be affected by serum or cell density. This phenomenon was also reported as having physiological relevance in kidney cells, in which both serum- and glucocorticoid-inducible kinase and NDRG2 were induced rapidly by aldosterone treatment.(25, 26)
In immunohistochemical study, hNDRG protein was abundantly expressed in the normal breast tissue but abruptly down-regulated in invasive breast cancer. Recently, it was reported that hNDRG2 expression in human meningeal tumor was regulated by hypermethylation.(27) Hypermethylation is a regional event that occurs frequently in GC-rich sequence, called CpG islands and often located within the 5' regulatory nontranscribed regions of genes. It has been recognized that aberrant hypermethylation of CpG islands in the promoters of certain tumor suppressor genes is known to be associated with transcriptional inactivation and loss of function and that promoter hypermethylation is often an early event in multistep carcinogenesis.(28) The author performed DNA methylation analysis of the hNDRG2 gene. In human tumor cell lines, MDA-MB-231 breast cancer cell line and SW620 colon cancer cell line showed both methylated and unmethylated DNA PCR products. However, unmethylated DNA of the NDRG2 gene was amplified in all 11 samples but methylated DNA was not amplified in any cases of the corresponding normal breast tissues. Also, hypermethylation was not found in all 11 tumorous samples examined. These findings suggest that hNDRG2 transcriptional gene regulation by hypermethylation may be tissue or organ specific event in in vivo tumorigenesis.
With the aid of univariate logistic analysis, hNDRG2 was significantly down-regulated in the invasive ductal carcinomatous tissue, compared to the normal ductal and lobular tissues. We propose that level of hNDRG2 protein may negatively contribute to conversion of the malignant phenotype in the tumorigenic process of the human invasive breast cancer. However, further investigation is needed to reveal the pathologic role and its importance of hNDRG2 protein in human breast carcinoma.
CONCLUSION
Our study showed that hNDRG2 was significantly down-regulated in the invasive ductal cancer tissue, and it was a correlation between overexpression of hNDRG2 and tumor size, histologic grade. Also hNDRG2 overexpression resulted in decrease of cell proliferation and regulation of genes (including cell signaling and cell cycle related genes) in MDA-MB-231 cell line.
These findings suggested that hNDRG2 can function, at least in part, as a regulator of cell cycle and tumor suppressor in the human invasive breast carcinoma. In addition, hNDRG2 protein indicates a prognostic tumor marker for human breast cancer.




 PDF
PDF ePub
ePub Citation
Citation Print
Print











 XML Download
XML Download