Abstract
Pseudomonas oryzihabitans is frequently found in various sites within hospital settings, including sink drains and respiratory therapy equipment. Although it rarely causes human infections, P. oryzihabitans has recently been considered a potential nosocomial pathogen, especially in immunocompromised hosts. We report our experience of an outbreak of P. oryzihabitans pseudobacteremia, presumably due to faulty aseptic preparation of a saline gauze canister.
Pseudomonas oryzihabitans is the current name for the organism previously called Chromobacterium typhiflavum and Flavimonas oryzihabitans [1]. It is a gram-negative and oxidase-negative, non-fermenting bacterium whose colonies are typically yellow-pigmented, rough or wrinkled after 48 h incubation on agar media [2, 3]. Although it remains a rare cause of human infections, P. oryzihabitans has more recently been considered a potential nosocomial pathogen, most often reported in immunocompromised hosts [4, 5, 6]. In the hospital setting, P. oryzihabitans is frequently found in various sites and has been recovered from sink drains and respiratory therapy equipment [1, 7]. P. oryzihabitans infections usually occur in cases involving catheters, with central venous catheter-related infections reported most frequently [8].
Pseudobacteremia describes apparent bacteremia that, after careful investigation, is generally clinically insignificant and typically due to a blood culture contaminant [9]. Accurate data about the incidence of pseudobacteremia are not known, but Weinstein et al. reported that up to 11% of nosocomial outbreaks were pseudoepidemics [10]. It is important to recognize and control pseudobacteremia outbreaks of these opportunistic pathogens because epidemics can be costly and time consuming and these pathogens can be a source of sepsis in at-risk patients. Here, we report our experience of a P. oryzihabitans pseudobacteremia outbreak, presumably arising from faulty aseptic preparation of a saline gauze canister.
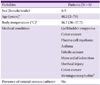
Between October 7 and October 11, 2011, 8 patients visited the emergency room of a tertiary hospital and were diagnosed with blood cultures positive for P. oryzihabitans. When these patients visited our emergency room (ER) complaining of febrile sensations in spite of ambiguous clinical signs and symptoms, blood cultures were ordered to test for systemic infections. Antimicrobial susceptibility testing was simultaneously performed using standard techniques. Blood samples were placed in culture bottles (Vital AER or Vital ANA, bioMérieux; Marcy-l'Etoile, France) and incubated for 24.48 h; P. oryzihabitans was identified in 10 samples from 8 patients using a Vitek system (bioMérieux). Antimicrobial susceptibility testing was also performed using the same system. All but 2 patients had a single positive blood culture result, and no microorganism was isolated from other sites. The characteristics of the affected patients, including demographic data, medical history, and presence of a central venous catheter, are shown in Table 1. None of the patients with blood cultures positive for P. oryzihabitans showed clinical signs of sepsis; this cluster of cultures positive for an unusual pathogen was suggestive of a pseudoepidemic. Review of microbiology laboratory records from the previous 12 months found no instances of P. oryzihabitans. During the same period, P. oryzihabitans was not isolated from clinical specimens from other wards. The ER environment was investigated to determine potential sources of infection and to implement control measures to stop the outbreak. Bacteriological cultures of ER environmental objects were performed.
A total of 22 environmental samples from the ER were cultured to test for a P. oryzihabitans cluster. Sites examined included the disinfectant, forceps, saline gauze cans, and gloves used for dressing wounds as well as hand-washing soap, tap water, tables, beds, chairs, and linens in the ER.
Among these environmental samples, only 1, from a saline gauze canister, was culture-positive for P. oryzihabitans. This culture result was compared to results of 16S rRNA sequencing of the same sample. Sequencing also revealed P. oryzihabitans. Antimicrobial susceptibility tests of the organism isolated from the saline gauze canister showed the same pattern as the patient samples. All isolates were sensitive to imipenem, gentamicin, penicillin, ciprofloxacin, ceftazidime, ampicillin, amoxicillin/clavulanic acid, trimethoprim/sulfamethoxazole, cefotaxime, and ceftazidime. All isolates were sensitive even to ampicillin, to which some Pseudomonas species have a known endogenous resistance. Saline-soaked cotton gauzes stored in canisters were used in our hospital to remove dirt from intact skin. However, these gauzes could have been used to clean disinfectant from the skin for access to veins and mistaken for cotton materials soaked in 70% ethanol, which were stored in similar canisters and were of the same color. Consequently, careful investigation of the above results revealed the outbreak to be pseudobacteremia; the most likely etiology was environmental contamination of the saline gauze can, which contaminated several blood cultures during collection of specimen.
After the outbreak, the infection control unit instituted infection control measures including removal of saline gauze cans from the ward, including the ER. After instituting control measures and monitoring blood culture results daily, no additional outbreaks have been documented in the ER. Therefore, we could conclude that we had found the focus of the P. oryzihabitans contamination and pseudobacteremia outbreak and that the outbreak was over.
Several reports have shown evidence of pseudobacteremia outbreaks caused by P. oryzihabitans. Eom et al. [11] reported an outbreak of P. oryzihabitans pseudobacteremia, presumably arising from faulty aseptic preparation of a blood culture set. In that report, genotypically identical isolates were identified based on pulsed-field gel electrophoresis (PFGE) patterns of 4 P. oryzihabitans isolates from patients with no signs or symptoms consistent with P. oryzihabitans bacteremia. Kim et al. [12] reported Enterobacter nimipressuralis pseudobacteremia from the use of contaminated saline-soaked cotton at the sites of venipuncture blood draw. Because Enterobacter clinical isolates could not be identified at the species level by 16S rRNA sequencing, the organism was accurately identified by sequencing of the hsp60 gene and infrequent-restriction-site polymerase chain reaction (IRS-PCR). In most reported instances of pseudobacteremia outbreaks, automated culture systems and molecular typing were used to find that hospital environmental contaminants, such as coagulase-negative cocci, Bacillus spp., Corynebacterium spp., Micrococcus spp., Propionibacterium spp., gram-positive bacilli, or Clostridium perfringens were the most common causes of outbreaks [13, 14]. Recently, Neulier et al. [15] reported a pseudo-outbreak of Pseudomonas putida respiratory infections. Investigation of the microbiology laboratory where 5 samples were processed by the same device found that P. putida isolates were obtained from a reusable container in this device. All isolates presented identical antibiotic susceptibility patterns; therefore, the authors declared this pseudo-outbreak of P. putida to be over without additional testing such as random amplified polymorphic DNA (RAPD) typing or PFGE. Similarly, we did not investigate the clonality of the environmental isolates in this study by 16S rRNA sequencing, RAPD typing, or PFGE. Although the clinical significance of the P. oryzihabitans outbreak presented here remains unclear, P. oryzihabitans is an infrequent cause of infection and was isolated from patients with no clinical signs of sepsis. In addition, antimicrobial susceptibility test results were the same for P. oryzihabitans isolated from both patients and the contamination source. We suggest that P. oryzihabitans from a contaminated saline gauze canister was an etiology of pseudobacteremia outbreak in our ER; however, molecular typing of the environmental and patient samples was not performed.
In conclusion, we found the focus of P. oryzihabitans contamination to be a saline gauze canister and could declare the pseudobacteremia outbreak of P. oryzihabitans to be over after daily monitoring of blood culture results. Our results confirm previous reports that pseudobacteremia is usually caused by hospital environmental contaminants and emphasize the importance of hospital surveillance to reduce outbreaks and protect other patients at risk.
Figures and Tables
References
1. Chaudhry HJ, Schoch PE, Cunha BA. Flavimonas oryzihabitans (CDC group Ve2). Infect Control Hosp Epidemiol. 1992; 13:485–488.
2. Sanchez-Carillo C, Blazquez R, Cernado E, Diaz MD, Bouza E. Infections due to Flavimonas oryzihabitans: a report of three cases. Clin Microbiol Newslett. 1996; 18:158–160.
3. Rahav G, Simhon A, Mattan Y, Moses AE, Sacks T. Infections with Chryseomonas luteola (CDC group Ve-1) and Flavimonas oryzihabitans (CDC group Ve-2). Medicine (Baltimore). 1995; 74:83–88.

4. Lam S, Isenberg HD, Edwards B, Hilton E. Community-acquired soft-tissue infections caused by Flavimonas oryzihabitans. Clin Infect Dis. 1994; 18:808–809.

5. Lin RD, Hsueh PR, Chang JC, Teng LJ, Chang SC, Ho SW, Hsieh WC, Luh KT. Flavimonas oryzihabitans bacteremia: clinical features andmicrobiological characteristics of isolates. Clin Infect Dis. 1997; 24:867–873.

6. Decker CF, Simon GL, Keiser JF. Flavimonas oryzihabitans (Pseudomonas oryzihabitans; CDC group Ve-2) bacteremia in the immunocompromised host. Arch Intern Med. 1991; 151:603–604.

7. Steinberg JP, Rio CD. Other gram-negative and gram-variable bacilli. In : Mandell GL, Bennett JE, Dolin R, editors. Principles and Practices of Infectious Diseases. 6th ed. New York: Churchill Livingstone;2005. p. 275168.
8. Marín M, Garca de Viedma D, Martn-Rabadn P, Rodrguez-Crixems M, Bouza E. Infection of hickman catheter by Pseudomonas (formerly Flavimonas) oryzihabitans traced to a synthetic bath sponge. J Clin Microbiol. 2000; 38:4577–4579.

11. Eom JS, Cheong HJ, Kim WJ. Pseudobacteremia outbreak of Pseudomonas oryzihabitans in an emergency department of a tertiary hospital in Korea. Infect Control Hosp Epidemiol. 2009; 30:803–804.

12. Kim DM, Jang SJ, Neupane GP, Jang MS, Kwon SH, Kim SW, Kim WY. Enterobacter nimipressuralis as a cause of pseudobacteremia. BMC Infect Dis. 2010; 10:315.
13. Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, Reller LB. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis. 1997; 24:584–602.





 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download