1. Hiramatsu K. Molecular evolution of MRSA. Microbiol Immunol. 1995; 39:531–543. PMID:
7494490.

2. Jevons MP. 'Celbenin'-resistant staphylococci. BMJ. 1961; 1:124–125.
3. Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant
Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A. 2002; 99:7687–7692. PMID:
12032344.
4. Hiramatsu K, Hanaki H, Ino T, Yabuta K, Oguri T, Tenover FC. Methicillin-resistant
Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J Antimicrob Chemother. 1997; 40:135–136. PMID:
9249217.
5. Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Downes FP, Shah S, Rudrik JT, Pupp GR, Brown WJ, Cardo D, Fridkin SK. Vancomycin-Resistant
Staphylococcus aureus Investigative Team. Infection with vancomycin-resistant
Staphylococcus aureus containing the vanA resistance gene. N Engl J Med. 2003; 348:1342–1347. PMID:
12672861.
6. Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant
Staphylococcus aureus in Western Australia. J Hosp Infect. 1993; 25:97–108. PMID:
7903093.
7. Riley TV, Pearman JW, Rouse IL. Changing epidemiology of methicillin-resistant
Staphylococcus aureus in Western Australia. Med J Aust. 1995; 163:412–414. PMID:
7476610.
8. Moreno F, Crisp C, Jorgensen JH, Patterson JE. Methicillin-resistant
Staphylococcus aureus as a community organism. Clin Infect Dis. 1995; 21:1308–1312. PMID:
8589164.
9. Aubry-Damon H, Legrand P, Brun-Buisson C, Astier A, Soussy CJ, Leclercq R. Reemergence of gentamicin-susceptible strains of methicillin-resistant
Staphylococcus aureus: roles of an infection control program and changes in aminoglycoside use. Clin Infect Dis. 1997; 25:647–653. PMID:
9314454.
10. Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002; 359:1819–1827. PMID:
12044378.

11. Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, Baba S, Fukui S, Lee JC, Hiramatsu K. Whole-genome sequencing of
Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J Bacteriol. 2005; 187:7292–7308. PMID:
16237012.
12. Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in
Staphylococcus aureus. Mol Microbiol. 1998; 29:527–543. PMID:
9720870.
13. Novick RP, Subedi A. The SaPIs: mobile pathogenicity islands of
Staphylococcus. Chem Immunol Allergy. 2007; 93:42–57. PMID:
17369699.
14. O'Neill AJ, Larsen AR, Skov R, Henriksen AS, Chopra I. Characterization of the epidemic European fusidic acid-resistant impetigo clone of
Staphylococcus aureus. J Clin Microbiol. 2007; 45:1505–1510. PMID:
17344365.
15. Ubeda C, Maiques E, Tormo MA, Campoy S, Lasa I, Barbé J, Novick RP, Penadés JR. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol Microbiol. 2007; 65:41–50. PMID:
17581119.
16. Tormo MA, Ferrer MD, Maiques E, Ubeda C, Selva L, Lasa I, Calvete JJ, Novick RP, Penadés JR.
Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J Bacteriol. 2008; 190:2434–2440. PMID:
18223072.
17. Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009; 323:139–141. PMID:
19119236.

18. Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. Complete genome sequence determination of a
Macrococcus caseolyticus strain JSCS5402 reflecting the ancestral genome of the human pathogenic staphylococci. J Bacteriol. 2009; 191:1180–1190. PMID:
19074389.
19. Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant
Staphylococcus aureus strain and a biofilm-producing methicillin-resistant
Staphylococcus epidermidis strain. J Bacteriol. 2005; 187:2426–2438. PMID:
15774886.
20. Lina G, Bohach GA, Nair SP, Hiramatsu K, Jouvin-Marche E, Mariuzza R. International Nomenclature Committee for Staphylococcal Superantigens. Standard nomenclature for the superantigens expressed by
Staphylococcus. J Infect Dis. 2004; 189:2334–2336. PMID:
15181583.
21. Langley R, Wines B, Willoughby N, Basu I, Proft T, Fraser JD. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-Fc alpha RI binding and serum killing of bacteria. J Immunol. 2005; 174:2926–2933. PMID:
15728504.
22. Baba T, Takeuchi F, Kuroda M, Ito T, Yuzawa H, Hiramatsu K. The Staphylococcus aureus genome. In : Ala'Aldeen D, Hiramatsu K, editors. Staphylococcus aureus Molecular and Clinical Aspects. Chichester, UK: Horwood Publishing;2004. p. 66–153.
23. Yamaguchi T, Nishifuji K, Sasaki M, Fudaba Y, Aepfelbacher M, Takata T, Ohara M, Komatsuzawa H, Amagai M, Sugai M. Identification of the
Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect Immun. 2002; 70:5835–5845. PMID:
12228315.
24. Manna AC, Cheung AL.
sarU, a
sarA homolog, is repressed by SarT and regulates virulence genes in
Staphylococcus aureus. Infect Immun. 2003; 71:343–353. PMID:
12496184.
25. Jamaluddin TZ, Kuwahara-Arai K, Hisata K, Terasawa M, Cui L, Baba T, Sotozono C, Kinoshita S, Ito T, Hiramatsu K. Extreme genetic diversity of methicillin-resistant
Staphylococcus epidermidis strains disseminated among healthy Japanese children. J Clin Microbiol. 2008; 46:3778–3783. PMID:
18832123.
26. Watanabe S, Ito T, Morimoto Y, Takeuchi F, Hiramatsu K. Precise excision and self-integration of a composite transposon as a model for spontaneous large-scale chromosome inversion/deletion of the
Staphylococcus haemolyticus clinical strain JCSC1435. J Bacteriol. 2007; 189:2921–2925. PMID:
17237177.
27. Gustafson J, Strässle A, Hächler H, Kayser FH, Berger-Bächi B. The
femC locus of
Staphylococcus aureus required for methicillin resistance includes the glutamine synthetase operon. J Bacteriol. 1994; 176:1460–1467. PMID:
7509336.
28. Maki H, Murakami K. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant
Staphylococcus aureus. J Bacteriol. 1997; 179:6944–6948. PMID:
9371438.
29. Hiramatsu K. Vancomycin resistance in staphylococci. Drug Resist Updat. 1998; 1:135–150. PMID:
16904400.

30. Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial
Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect Immun. 2004; 72:1210–1215. PMID:
14742578.
31. Rosenstein R, Nerz C, Biswas L, Resch A, Raddatz G, Schuster SC, Gätz F. Genome analysis of the meat starter culture bacterium
Staphylococcus carnosus TM300. Appl Environ Microbiol. 2009; 75:811–822. PMID:
19060169.
32. Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant
Staphylococcus aureus N315. Antimicrob Agents Chemother. 1999; 43:1449–1458. PMID:
10348769.
33. Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome
mec, encodes methicillin resistance in
Staphylococcus aureus. Antimicrob Agents Chemother. 2000; 44:1549–1555. PMID:
10817707.
34. Sjöström JE, Löfdahl S, Philipson L. Transformation reveals a chromosomal locus of the gene(s) for methicillin resistance in
Staphylococcus aureus. J Bacteriol. 1975; 123:905–915. PMID:
125746.
35. Kuhl SA, Pattee PA, Baldwin JN. Chromosomal map location of the methicillin resistance determinant in
Staphylococcus aureus. J Bacteriol. 1978; 135:460–465. PMID:
249312.
36. Stewart GC, Rosenblum ED. Genetic behavior of the methicillin resistance determinant in
Staphylococcus aureus. J Bacteriol. 1980; 144:1200–1202. PMID:
6254946.
37. Brown DF, Reynolds PE. Intrinsic resistance to beta-lactam antibiotics in
Staphylococcus aureus. FEBS Lett. 1980; 122:275–278. PMID:
7202719.
38. Hartman BJ, Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in
Staphylococcus aureus. J Bacteriol. 1984; 158:513–516. PMID:
6563036.
39. Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant
Staphylococcus aureus. Antimicrob Agents Chemother. 1985; 28:397–403. PMID:
3878127.
40. Ubukata K, Yamashita N, Konno M. Occurrence of a beta-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1985; 27:851–857. PMID:
3848294.

41. Matsuhashi M, Song MD, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in
Staphylococcus aureus. J Bacteriol. 1986; 167:975–980. PMID:
3638304.
42. Song MD, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant
Staphylococcus aureus by gene fusion. FEBS Lett. 1987; 221:167–171. PMID:
3305073.
43. Tesch W, Ryffel C, Strässle A, Kayser FH, Berger-Bächi B. Evidence of a novel staphylococcal
mec-encoded element (
mecR) controlling expression of penicillin-binding protein 2'. Antimicrob Agents Chemother. 1990; 34:1703–1706. PMID:
2285282.
44. Hürlimann-Dalel RL, Ryffel C, Kayser FH, Berger-Bächi B. Survey of the methicillin resistance-associated genes
mecA,
mecR1-mecI, and
femA-femB in clinical isolates of methicillin-resistant
Staphylococcus aureus. Antimicrob Agents Chemother. 1992; 36:2617–2621. PMID:
1362343.
45. Suzuki E, Kuwahara-Arai K, Richardson JF, Hiramatsu K. Distribution of
mec regulator genes in methicillin-resistant
Staphylococcus clinical strains. Antimicrob Agents Chemother. 1993; 37:1219–1226. PMID:
8328773.
46. Ryffel C, Kayser FH, Berger-Bächi B. Correlation between regulation of
mecA transcription and expression of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 1992; 36:25–31. PMID:
1375449.
47. Hiramatsu K, Asada K, Suzuki E, Okonogi K, Yokota T. Molecular cloning and nucleotide sequence determination of the regulator region of
mecA gene in methicillin-resistant
Staphylococcus aureus (MRSA). FEBS Lett. 1992; 298:133–136. PMID:
1544435.
48. Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suzuki E, Hiramatsu K. Suppression of methicillin resistance in a
mecA-containing pre-methicillin-resistant
Staphylococcus aureus strain is caused by the
mecI-mediated repression of PBP 2' production. Antimicrob Agents Chemother. 1996; 40:2680–2685. PMID:
9124822.
49. Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, Hiramatsu K. Structural comparison of three types of staphylococcal cassette chromosome
mec integrated in the chromosome in methicillin-resistant
Staphylococcus aureus. Antimicrob Agents Chemother. 2001; 45:1323–1336. PMID:
11302791.
50. Hiramatsu K, Kondo N, Ito T. Genetic basis for molecular epidemiology of MRSA. J Infect Chemother. 1996; 2:117–129.

51. Stokes HW, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989; 3:1669–1683. PMID:
2560119.

52. Hiramatsu K, Katayama Y, Yuzawa H, Ito T. Molecular genetics of methicillin-resistant
Staphylococcus aureus. Int J Med Microbiol. 2002; 292:67–74. PMID:
12195737.
53. Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K. Staphylococcal cassette chromosome
mec (
SCCmec) typing of methicillin-resistant
Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for
SCCmec elements. Antimicrob Agents Chemother. 2006; 50:1001–1012. PMID:
16495263.
54. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. Combination of multiplex PCRs for staphylococcal cassette chromosome
mec type assignment: rapid identification system for
mec,
ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007; 51:264–274. PMID:
17043114.
55. Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, Daum RS, Hiramatsu K. Novel type of staphylococcal cassette chromosome
mec identified in community-acquired methicillin-resistant
Staphylococcus aureus strains. Antimicrob Agents Chemother. 2002; 46:1147–1152. PMID:
11897611.
56. Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel type V staphylococcal cassette chromosome
mec driven by a novel cassette chromosome recombinase,
ccrC. Antimicrob Agents Chemother. 2004; 48:2637–2651. PMID:
15215121.
57. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome
mec (SCC
mec): guidelines for reporting novel SCC
mec elements. Antimicrob Agents Chemother. 2009; 53:4961–4967. PMID:
19721075.
58. Okuma K, Iwakawa K, Turnidge JD, Grubb WB, Bell JM, O'Brien FG, Coombs GW, Pearman JW, Tenover FC, Kapi M, Tiensasitorn C, Ito T, Hiramatsu K. Dissemination of new methicillin-resistant
Staphylococcus aureus clones in the community. J Clin Microbiol. 2002; 40:4289–4294. PMID:
12409412.
59. Hisata K, Kuwahara-Arai K, Yamanoto M, Ito T, Nakatomi Y, Cui L, Baba T, Terasawa M, Sotozono C, Kinoshita S, Yamashiro Y, Hiramatsu K. Dissemination of methicillin-resistant staphylococci among healthy Japanese children. J Clin Microbiol. 2005; 43:3364–3372. PMID:
16000461.

60. Ruppé E, Barbier F, Mesli Y, Maiga A, Cojocaru R, Benkhalfat M, Benchouk S, Hassaine H, Maiga I, Diallo A, Koumaré AK, Ouattara K, Soumaré S, Dufourcq JB, Nareth C, Sarthou JL, Andremont A, Ruimy R. Diversity of staphylococcal cassette chromosome
mec structures in methicillin-resistant
Staphylococcus epidermidis and
Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob Agents Chemother. 2009; 53:442–449. PMID:
19001111.
61. Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant
Staphylococcus aureus. Trends Microbiol. 2001; 9:486–493. PMID:
11597450.
62. Chambers HF. The changing epidemiology of
Staphylococcus aureus? Emerg Infect Dis. 2001; 7:178–182. PMID:
11294701.
63. Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF. A proteolytic transmembrane signaling pathway and resistance to beta-lactams in staphylococci. Science. 2001; 291:1962–1965. PMID:
11239156.
64. Hiramatsu K, Suzuki E, Takayama H, Katayama Y, Yokota T. Role of penicillinase plasmids in the stability of the
mecA gene in methicillin-resistant
Staphylococcus aureus. Antimicrob Agents Chemother. 1990; 34:600–604. PMID:
2344167.
65. Wada A, Katayama Y, Hiramatsu K, Yokota T. Southern hybridization analysis of the
mecA deletion from methicillin-resistant
Staphylococcus aureus. Biochem Biophys Res Commun. 1991; 176:1319–1325. PMID:
2039513.
66. Lee SM, Ender M, Adhikari R, Smith JM, Berger-Bächi B, Cook GM. Fitness cost of staphylococcal cassette chromosome
mec in methicillin-resistant
Staphylococcus aureus by way of continuous culture. Antimicrob Agents Chemother. 2007; 51:1497–1499. PMID:
17283194.
67. Chatterjee SS, Chen L, Joo HS, Cheung GY, Kreiswirth BN, Otto M. Distribution and regulation of the mobile genetic element-encoded phenol-soluble modulin PSM-mec in methicillin-resistant
Staphylococcus aureus. PLoS One. 2011; 6:e28781. PMID:
22174895.
68. Kaito C, Sekimizu K. Colony spreading in
Staphylococcus aureus. J Bacteriol. 2007; 189:2553–2557. PMID:
17194792.
69. Kaito C, Omae Y, Matsumoto Y, Nagata M, Yamaguchi H, Aoto T, Ito T, Hiramatsu K, Sekimizu K. A novel gene,
fudoh, in the SCC
mec region suppresses the colony spreading ability and virulence of
Staphylococcus aureus. PLoS ONE. 2008; 3:e3921. PMID:
19079549.
70. Hongo I, Baba T, Oishi K, Morimoto Y, Ito T, Hiramatsu K. Phenol-soluble modulin alpha 3 enhances the human neutrophil lysis mediated by Panton-Valentine leukocidin. J Infect Dis. 2009; 200:715–723. PMID:
19653829.
71. Kaito C, Saito Y, Nagano G, Ikuo M, Omae Y, Hanada Y, Han X, Kuwahara-Arai K, Hishinuma T, Baba T, Ito T, Hiramatsu K, Sekimizu K. Transcription and translation products of the cytolysin gene
psm-mec on the mobile genetic element SCC
mec regulate
Staphylococcus aureus virulence. PLoS Pathog. 2011; 7:e1001267. PMID:
21304931.
72. Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, Hasegawa S, Yamaguchi H, Inokuchi K, Ito T, Hiramatsu K, Sekimizu K. Mobile genetic element SCC
mec-encoded
psm-mec RNA suppresses translation of
agrA and attenuates MRSA virulence. PLoS Pathog. 2013; 9:e1003269. PMID:
23592990.
73. Luong TT, Ouyang S, Bush K, Lee CY. Type 1 capsule genes of
Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J Bacteriol. 2002; 184:3623–3629. PMID:
12057957.
74. Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, Foster TJ, Moore CE, Hurst L, Atkin R, Barron A, Bason N, Bentley SD, Chillingworth C, Chillingworth T, Churcher C, Clark L, Corton C, Cronin A, Doggett J, Dowd L, Feltwell T, Hance Z, Harris B, Hauser H, Holroyd S, Jagels K, James KD, Lennard N, Line A, Mayes R, Moule S, Mungall K, Ormond D, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Sharp S, Simmonds M, Stevens K, Whitehead S, Barrell BG, Spratt BG, Parkhill J. Complete genomes of two clinical
Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci U S A. 2004; 101:9786–9791. PMID:
15213324.
75. Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. Genome-based analysis of virulence genes in a non-biofilm-forming
Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol. 2003; 49:1577–1593. PMID:
12950922.
76. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant
Staphylococcus aureus. Lancet. 2006; 367:731–739. PMID:
16517273.
77. Wu S, Piscitelli C, de Lencastre H, Tomasz A. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of
mecA from a methicillin susceptible strain of
Staphylococcus sciuri. Microb Drug Resist. 1996; 2:435–441. PMID:
9158816.
78. Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. 2010; 54:4352–4359. PMID:
20679504.

79. Hiramatsu K, Tsubakishita S, Matsuo M, Sasaki T. Molecular Evolution of MRSA 2010. In : 12th Western Pacific Congress on Chemotherapy and Infectious Diseases; 2011 Dec 2-5; Singapore. p. 11–18.
80. Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. Staphylococcal cassette chromosome
mec-like element in
Macrococcus caseolyticus. Antimicrob Agents Chemother. 2010; 54:1469–1475. PMID:
20086147.
81. García-Álvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. Meticillin-resistant
Staphylococcus aureus with a novel
mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011; 11:595–603. PMID:
21641281.
82. Shore AC, Deasy EC, Slickers P, Brennan G, O'Connell B, Monecke S, Ehricht R, Coleman DC. Detection of staphylococcal cassette chromosome
mec type XI carrying highly divergent
mecA,
mecI,
mecR1,
blaZ, and
ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant
Staphylococcus aureus. Antimicrob Agents Chemother. 2011; 55:3765–3773. PMID:
21636525.
83. Harrison EM, Paterson GK, Holden MT, Morgan FJ, Larsen AR, Petersen A, Leroy S, De Vliegher S, Perreten V, Fox LK, Lam TJ, Sampimon OC, Zadoks RN, Peacock SJ, Parkhill J, Holmes MA. A
Staphylococcus xylosus isolate with a new
mecC allotype. Antimicrob Agents Chemother. 2013; 57:1524–1528. PMID:
23274660.
84. Schnellmann C, Gerber V, Rossano A, Jaquier V, Panchaud Y, Doherr MG, Thomann A, Straub R, Perreten V. Presence of new
mecA and
mph(C) variants conferring antibiotic resistance in
Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J Clin Microbiol. 2006; 44:4444–4454. PMID:
17005735.
85. Fontana R, Ligozzi M, Pittaluga F, Satta G. Intrinsic penicillin resistance in enterococci. Microb Drug Resist. 1996; 2:209–213. PMID:
9158761.

86. Williamson R, le Bouguénec C, Gutmann L, Horaud T. One or two low affinity penicillin-binding proteins may be responsible for the range of susceptibility of
Enterococcus faecium to benzylpenicillin. J Gen Microbiol. 1985; 131:1933–1940. PMID:
3850924.
87. el Kharroubi A, Jacques P, Piras G, Van Beeumen J, Coyette J, Ghuysen JM. The
Enterococcus hirae R40 penicillin-binding protein 5 and the methicillin-resistant
Staphylococcus aureus penicillin-binding protein 2' are similar. Biochem J. 1991; 280:463–469. PMID:
1747121.
88. Piras G, Raze D, el Kharroubi A, Hastir D, Englebert S, Coyette J, Ghuysen JM. Cloning and sequencing of the low-affinity penicillin-binding protein 3r-encoding gene of
Enterococcus hirae S185: modular design and structural organization of the protein. J Bacteriol. 1993; 175:2844–2852. PMID:
8491705.
89. Mongkolrattanothai K, Boyle S, Murphy TV, Daum RS. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing
pbp4 and
tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in
Staphylococcus aureus. Antimicrob Agents Chemother. 2004; 48:1823–1836. PMID:
15105141.
90. Wyke AW, Ward JB, Hayes MV, Curtis NA. A role
in vivo for penicillin-binding protein-4 of
Staphylococcus aureus. Eur J Biochem. 1981; 119:389–393. PMID:
7308191.
91. Henze UU, Roos M, Berger-Bächi B. Effects of penicillin-binding protein 4 overproduction in
Staphylococcus aureus. Microb Drug Resist. 1996; 2:193–199. PMID:
9158759.
92. Yamakawa J, Aminaka M, Okuzumi K, Kobayashi H, Katayama Y, Kondo S, Nakamura A, Oguri T, Hori S, Cui L, Ito T, Jin J, Kurosawa H, Kaneko K, Hiramatsu K. Heterogeneously vancomycin-intermediate
Staphylococcus aureus (hVISA) emerged before the clinical introduction of vancomycin in Japan: a retrospective study. J Infect Chemother. 2012; 18:406–409. PMID:
22033576.
93. Ryffel C, Strässle A, Kayser FH, Berger-Bächi B. Mechanisms of heteroresistance in methicillin-resistant
Staphylococcus aureus. Antimicrob Agents Chemother. 1994; 38:724–728. PMID:
8031036.
94. Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in
Staphylococcus aureus. Mol Microbiol. 2003; 49:807–821. PMID:
12864861.
95. Kuroda M, Kuwahara-Arai K, Hiramatsu K. Identification of the up- and down-regulated genes in vancomycin-resistant
Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem Biophys Res Commun. 2000; 269:485–490. PMID:
10708580.
96. Aiba Y, Katayama Y, Hishinuma T, Murakami-Kuroda H, Cui L, Hiramatsu K. Mutation of RNA polymerase β-subunit gene promotes hetero-to-homo conversion of β-lactam resistance of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. [Submitted].
97. Kondo N, Kuwahara-Arai K, Kuroda-Murakami H, Tateda-Suzuki E, Hiramatsu K. Eagle-type methicillin resistance: new phenotype of high methicillin resistance under mec regulator gene control. Antimicrob Agents Chemother. 2001; 45:815–824. PMID:
11181367.
98. Matsuo M, Hishinuma T, Katayama Y, Cui L, Kapi M, Hiramatsu K. Mutation of RNA polymerase beta subunit (rpoB) promotes hVISA-to-VISA phenotypic conversion of strain Mu3. Antimicrob Agents Chemother. 2011; 55:4188–4195. PMID:
21746940.
99. Hiramatsu K, Igarashi M, Morimoto Y, Baba T, Umekita M, Akamatsu Y. Curing bacteria of antibiotic resistance: reverse antibiotics, a novel class of antibiotics in nature. Int J Antimicrob Agents. 2012; 39:478–485. PMID:
22534508.

100. Uchiyama I. MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucleic Acids Res. 2007; 35:D343–D346. PMID:
17135196.

101. Uchiyama I. Hierarchical clustering algorithm for comprehensive orthologous-domain classification in multiple genomes. Nucleic Acids Res. 2006; 34:647–658. PMID:
16436801.

102. Clarke SR, Harris LG, Richards RG, Foster SJ. Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of
Staphylococcus aureus. Infect Immun. 2002; 70:6680–6687. PMID:
12438342.
103. Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in
Staphylococcus aureus. PLoS One. 2007; 2:e1120. PMID:
17971880.
104. Uchiyama I. Multiple genome alignment for identifying the core structure among moderately related microbial genomes. BMC Genomics. 2008; 9:515. PMID:
18976470.

105. Yarwood JM, McCormick JK, Paustian ML, Orwin PM, Kapur V, Schlievert PM. Characterization and expression analysis of
Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J Biol Chem. 2002; 277:13138–13147. PMID:
11821418.
106. Highlander SK, Hultén KG, Qin X, Jiang H, Yerrapragada S, Mason EO Jr, Shang Y, Williams TM, Fortunov RM, Liu Y, Igboeli O, Petrosino J, Tirumalai M, Uzman A, Fox GE, Cardenas AM, Muzny DM, Hemphill L, Ding Y, Dugan S, Blyth PR, Buhay CJ, Dinh HH, Hawes AC, Holder M, Kovar CL, Lee SL, Liu W, Nazareth LV, Wang Q, Zhou J, Kaplan SL, Weinstock GM. Subtle genetic changes enhance virulence of methicillin resistant and sensitive
Staphylococcus aureus. BMC Microbiol. 2007; 7:99. PMID:
17986343.

107. Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, Meaney WJ, Smyth CJ. Characterization of a putative pathogenicity island from bovine
Staphylococcus aureus encoding multiple superantigens. J Bacteriol. 2001; 183:63–70. PMID:
11114901.
108. Ubeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, Penadés JR. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile
Staphylococcus aureus pathogenicity islands. Mol Microbiol. 2003; 49:193–210. PMID:
12823821.
109. Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant
Staphylococcus aureus. Lancet. 2001; 357:1225–1240. PMID:
11418146.
110. Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of
Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008; 190:300–310. PMID:
17951380.
111. Subedi A, Ubeda C, Adhikari RP, Penadés JR, Novick RP. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiology. 2007; 153:3235–3245. PMID:
17906123.

112. Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27
Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005; 102:5174–5179. PMID:
15788529.
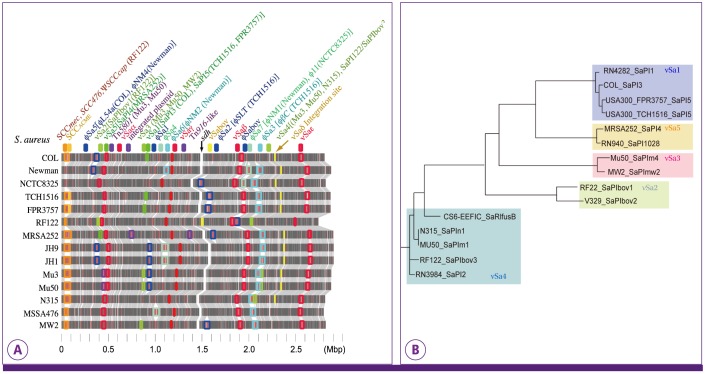
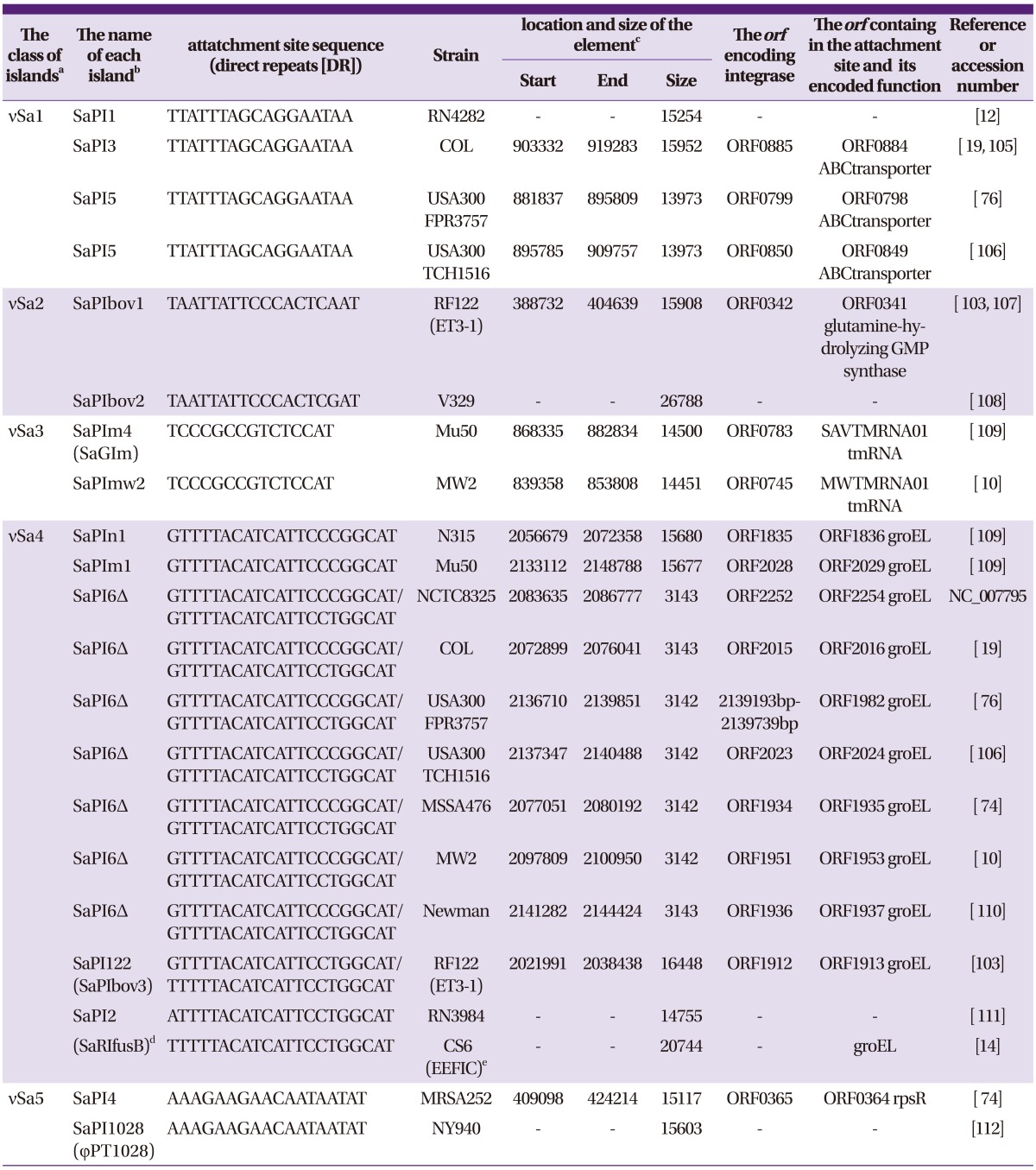
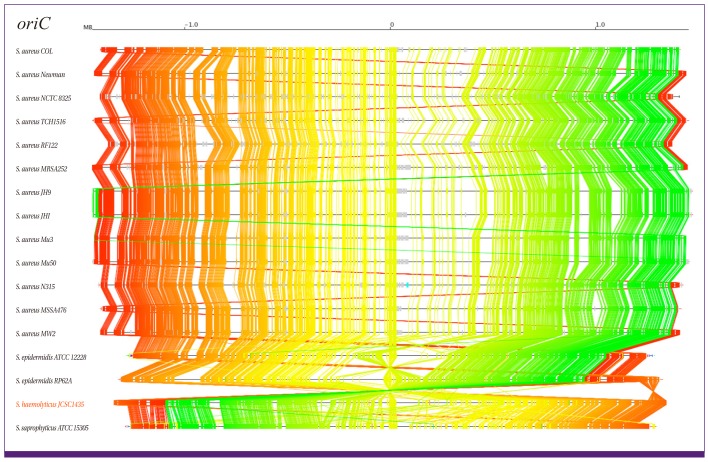
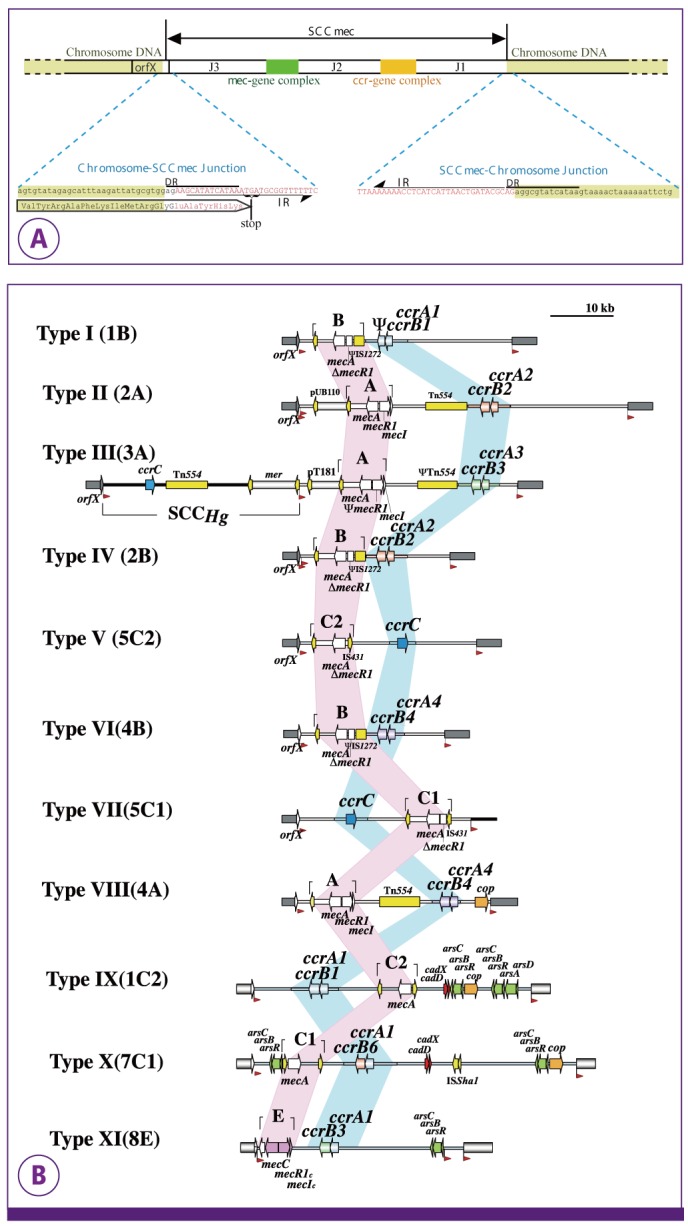




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download