Abstract
Influenza is a viral infection of the respiratory tract. Infection is normally confined to the upper respiratory tract but certain viral strains have evolved the ability to infect the lower respiratory tract, including the alveoli, leading to inflammation and a disease pattern of diffuse alveolar damage. Factors leading to this sequence of events are novel influenza strains, or strains that have viral proteins, in particular the NS1 protein that allow it to escape the innate immune system. There are three main barriers that prevent infection of pneumocytes - mucin, host defence lectins and cells such as macrophages. Viruses have developed strategies such as neuraminidase and glycosylation patterns that allow this evasion. Though there has been much investment in antiviral drugs, it is proposed that more attention should be directed towards developing or utilizing compounds that enhance the ability of the innate immune system to combat viral infection.
1918 was not a good year. Europe was not only in the final throes of a war that had extracted a heavy economic and physical burden on society with a tremendous loss of life. There was also the emergence of a new form of respiratory infection that was resulting in a 2-8% mortality. Doctors in Europe and the United States were battling to comprehend this emerging infection that had spread around military barracks and into the population with a transmission from humans to humans that had not been seen before. Researchers in the United States and United Kingdom examined the lungs of young soldiers who had died within a few days of contacting the disease. They found that the changes in the lungs were typified by the appearance of "... a non-cellular homogeneous exudate which was eosinophilic, and this was thought to be due to the presence of albumin coming from the serum..." The only comparison that this disease could be compared with was in the poisoning due to chlorine gas [1].
We now know that the agent that produced the disease was the influenza virus (most likely of avian origin) [2, 3] and that the changes seen were representative of a picture of diffuse alveolar damage [4].
Even after the discovery of the influenza virus there was not much interest from a pathological point of view on how the virus led to death. During the H2N2 and H3N2 pandemics of the 1950s and 1960s, limited autopsy and biopsy pathology indicated that bronchitis was the more common presentation and that a primary viral pneumonia was mainly seen in young patients, often with pre-existing disease such as mitral stenosis [5, 6]. Vaccination and intensive care with artificial ventilation was not readily available, so the overall picture was that of younger patients (possibly without prior virus exposure) succumbing to a fatal viral pneumonia within a number of days, with older subjects only developing bronchitis. Even with the advent of immunohistochemistry to identify viral antigen in biopsy and autopsy tissue, only a necrotizing bronchitis was seen and antigen was not normally present in the alveoli [7].
The emergence of H5N1 in isolated cases and the 2009 pandemic demonstrated a crucial factor in the understanding of influenza pathogenesis. In fatal case in the 2009 pandemic large numbers of influenza antigen positive cells were present in the lungs of fatal cases [8]. In contrast, patients who died of H5N1 rarely had abundant numbers of antigen positive cells - even though changes of diffuse alveolar damage (DAD) were present [9-11]. Since this latter group of patients died after antiviral and intensive care therapy [12-14], the picture was similar to that seen in SARS, where there was an acute viral induced disease with antigen present followed by a second phase of DAD where antigen was not identified but an evolving process of alveolar destruction followed by repair had commenced [15].
From the discussion above it becomes clear that the severe pulmonary burden occurs when the body is exposed to a viral agent to which there is no pre-existing immunity and in which most active viral replication occurs in the first 7 days followed by a prolonged virus-free exudative phase. This pattern appears similar in many viral pneumonic pictures and is orchestrated by the actions of the innate immune defence system [16].
The interaction of influenza with the innate immune defence system mechanism occurs broadly in 2 phases - the first is a circumvention of the natural extracellular host barriers and the second is a hijacking of the intracellular pathways by the virus in order to prevent an antiviral response and direct the cellular machinery towards replication of viral progeny. There are three barriers that influenza needs to defeat in order to successfully infect epithelial cells - mucin, host defence lectins and alveolar macrophages.
The key factors of the influenza virus that allow it to escape the natural defence mechanism are the surface protein haemagglutinin (HA) and neuraminidase (NA). The first protein is called a haemagglutinin because studies by Hirst in 1942 demonstrated that the influenza virus was able to cause haemagglutination of red blood cells [17]. Further studies by Blixt and colleagues identified the component on the red blood cells that interacted with the HA and this was called sialic acid (Sia).
Sia is the name given to the nonulosonic carbohydrate neuraminic acid (Neu) that contains an acetyl group at the fifth carbon position, hence Neu5Ac. In many animal species the acetyl group also undergoes enzymatic transformation to form a glycolyl group - Neu5Gc. Humans lack the enzyme that performs this transformation, however small amounts of Neu5Gc may be incorporated into humans from the diet [18, 19].
The second key factor that allows viral infection is the presence of the NA enzyme that is able to cleave the Sia from an adjacent sugar (often galactose or Gal). This viral NA, however does not cleave all Sia equally and most strains have a preference for cleaving the Sia when it is linked to the Gal in an α2-3 configuration (so called Sia α2-3Gal)(reviewed by Air [20]). Once inside the cell the internal genes and proteins come into play and play a more dominant role than HA and NA.
Two seminal studies from the 1980s by Paulson and colleagues demonstrated key determinants about the Sia-HA interaction. The first was that viruses isolated from different animal species differed in their ability to cause haemagglutination [21]. Viruses isolated from humans and swine preferably agglutinated cells that contained the Sia-Gal in an α2-6 linkage, but when viruses from horses and avian sources were used, they preferred agglutination when the linkage was α2-3. The second key finding was the use of plant lectins that were discriminatory in their binding to different Sia-Gal interactions [22]. The lectin from the Elderberry bush, Sambuccus nigra(SNA), bound Siaα2-6Gal, whereas the lectin from the bark of Maackia amurensis (MAA) preferentially bound glycans with Siaα2-3Gal. These lectins were used in binding to the human respiratory tract and demonstrated that ciliated cells - the targets of influenza - bound mainly SNA, indicating an abundance of Siaα2-6, but goblet cells bound mainly MAA, indicating mainly α2-3. Putting these 2 findings together, it was hypothesized that avian viruses (mainly agglutinating red blood cells with Siaα2-3Gal) would not readily infect humans as the target cells were mainly Siaα2-6 (as demonstrated by SNA binding). In the 1980's Scholtissek proposed that swine could act as an intermediate host [23], and lectin binding by Ito and colleagues inferred that the swine respiratory tract had both binding with SNA and MAA, thus potentially acting as a mixing vessel [24]. Recent lectin binding studies [25, 26] and ex-vivo infections by van Poucke and colleagues, however, have shown that there may be more variation in the swine respiratory tract than was previously demonstrated [27].
The mucus that is present in the airways is a mixture of cells, cellular debris and polypetpides held together by macromolecular consituents called mucins that are either in the fluid or on the surface of cells. Mucin is not a homogeneous protein but a family of glycoproteins that may either be secreted (such as MUC5AC and MUC5B) or membrane associated, that may function as cellular receptors (MUC1, MUC4, MUC11, MUC13, MUC15 and MUC20). There are a number of excellent reviews on the different types of these mucins available [28, 29]. The first set of mucins are normally secreted by goblet cells of the surface epithelium (MUC5AC and MUC5B) and submucous glands (MUC5B) present in the trachea and bronchi, and the membrane associated mucins, such as MUC1 and MUC4, are present at the apical surface of ciliated cells (Fig. 1). The effect of mucin on influenza virus replication was initially investigated in 1942 [30]. The lectin binding studies by Baum and Paulson [22] and Lo-Guidice and colleagues [31] suggested that mucin was mainly α2-3 sialylated, and this is in keeping with mass spectrographic data that has determined the O-glycan components of mucin (Nicholls et el, unpublished results). Mucins therefore may trap avian influenza viruses because these have an affinity for α2-3 [32]. However, the successful ability of influenza to infect the respiratory epithelium is most likely due to the preference of the viral NA to cleave α2-3 linkages compared with α2-6 linkages. Because the mucin is highly sialylated, the acidic nature of the Sia results in entrapment of water giving a viscous gel like quality to the mucin that acts as a physical barrier to infection. As children have fewer numbers of submucous glands and goblet cells than adults [33], this reduction in number and quantity may be more susceptible to avain virus infection, such as H5N1 compared with the adult population.
The functional unit of the lung is the alveolus and this is involved in gas exchange. To achieve this function requires as narrow a thickness as possible for efficient gas exchange, thus the presence of mucin in this environment would be a disadvantage as it would create a barrier to gas exchange. The body, however, has another line of defence to replace mucin and there are compounds called lectins which are proteins that bind to specific carbohydrate structures (reviewed by Ng and colleagues [34]). As the influenza HA and NA are glycoproteins, they undergo a process called glycosylation which means that glycans are added to the Asn residue of the sequence Asn-X-Ser/Thr, where X can be any amino acid except proline. In the normal glycosylation pathway the N-glycans are capped with Sia, however owing to the action of the viral NA, influenza itself is not sialylated but has an exposed galactose [35]. The glycosylation of HA is important in the development of antigenic variation but also plays a role in recognition by the host lectin mechanism [36]. In 2007 Reading and colleagues showed that the degree of glycosylation affected the ability of certain viruses to replicate in the respiratory tract of mice [37]. Viruses that lacked glycosylation of HA were able to cause more disease in mice than viruses that were glycosylated.
Three classes of lectin have been demonstrated to show anti-influenza activity - the C-type lectins, S-type lectins and pentraxins (reviewed in Ng and colleagues [34]. The first type. C-lectins, are calcium dependent (C-type) and recognize mannose or galactose. The C-type lectins have a subgroup called collectins (because of a collagenous domain). The collectins have a trimer composed of polypeptides that have a carbohydrate recognition domain. In the lung the 2 main lectins involved in anti-influenza activity are surfactant proteins A and D. These 2 collectins have different mechanisms. SP-A is sialylated and so acts as a decoy receptor (so called γ-inhibition) - thus its ability to prevent influenza infection is not dependent on the degree of HA glycosylation [38]. In contrast SP-D binds mannose type oligosaccharides and has been demonstrated to interact with influenza virus HA involved in haemagglutination inhibition, and is also able to mediate aggregation of viral particles and inhibit HA activity through interaction of the viral HA or NA (Fig. 2). Since SP-D interacts with the mannose, its antiviral effect will be dependent on the degree of glycosylation present, thus the changes in HA associated with drift due to increasing glycosylation will lead to greater neutralization by SP-D and thus reduced virulence.
The influence of SP-D on the innate immune response has been studied in SP-D deficient mice, which showed enhanced viral replication of glycosylated influenza virus. This change can be rectified if there is overexpression of wild type SP-D proteins [39]. Reading and colleagues have further shown that elevated glucose can interact with SP-D resulting in more severe disease in diabetic mice [40]. This finding is of note when it was found that patients with diabetes or obesity were more prone to developing severe disease in the 2009 pandemic [41].
A third lectin that is involved in the innate immune response is mannose binding lectin (MBL) which has the added effect of interacting with the complement pathway leading to lysis or phagocytosis.
One of the histological features seen in patients with H5N1 has been the increased numbers of macrophages in the alveolar space [42]. Studies of the ex vivo tissues infected with H5N1 have shown positive antigen in these cells [43]. Unlike the different types of epithelial cells present in the bronchus, alveolar macrophages appear equally susceptible to human and avian influenza virus infection, though H5N1 shows less productive replication in alveolar macrophages than bone marrow derived macrophages [44, 45]. Macrophages have receptors for mannose and galactose (MMR and MGL) so this interaction can be Sia independent [46]. As influenza infection appears different in alveolar macrophages compared with bone marrow derived macrophages, extrapolation of studies using the latter to the real clinical picture should be interpreted with caution.
These are low in number in the alveoli of normal lungs but as a "first responder" will be recruited into areas of influenza infection. Experimental infection of neutrophils with H5N1 resulted in an increase in matrix gene but this was not found in infection with H1N1 (Chan RWY, Personal Communication). Compared with macrophages this cell type has not been investigated in as much detail but this requires more investigation.
Similar to neutrophils these cells do not constitute a major population of inflammatory cells within normal alveoli. They do however appear to be closely involved in later stages of the inflammatory response, either through direct activation using the interferon receptor pathway (see below) or through type I IFN-mediated trans-presentation of IL-15 by dendritic cells (reviewed by Hwang and colleagues [47]).
The two main cells present in the alveoli are the Type 1 and Type 2 pneumocytes. Type 1 pneumocytes are large and flattened and incapable of replication. The type 2 pneumocytes are smaller and responsible for surfactant production involved in the antiviral defence mentioned above (reviewed in Crouch and Hillaire [48, 49]. When type 1 cells are damaged the type 2 will differentiate into type 1 cells. The diffuse alveolar damage picture seen in the fatal cases of 1918 H1N1 and H5N1 is due to the destruction of the type 1 pneumocytes with a consequent disruption of the integrity of the vascular-epithelial interface leading to an outpouring of serum proteins including albumin - leading to the hyaline membranes.
It is therefore important to understand whether influenza virus targets mainly type 1 or type 2 epithelial cells. Van Riel and colleagues have performed virus binding assays and demonstrated that H5N1 bound to type 2 cells within the lung with the type 1 cells binding seasonal H1N1 [50]. This has been corroborated by ex vivo infections of lung tissues that demonstrated viral egress from type 2 rather than type 1 cells [51]. This implies that the destruction of type 1 cells leading to diffuse alveolar damage might not be due to a direct viral cytopathic effect but may be secondary to a cytokine mediated destruction. Analysis of this effect in the laboratory is challenging as keeping isolated type 2 cells in their native state is difficult as they rapidly differentiate into type 1-like cells [44]. Though many investigators have used the A549 cells as "representing" the lung epithelium, this cell line is derived from a type 2 pneumoctye tumour and does not faithfully represent their non-neoplastic counterpart (e.g lack surfactant production and require trypsin to be added for efficient virus replication).
The emergence of H5N1 in 1997 caused a great deal of concern as compared with seasonal influenza infection there was a significantly high morbidity and mortality. The question for clinicians and researchers was whether this was just due to properties of a "new" virus to which there was little human immunity or whether there were intrinsic viral properties that resulted in a high mortality. Infection of macrophages or epithelial cells in the in vitro setting pointed to the latter scenario [52, 53], and transcription analysis showed that H5N1 was more potent than seasonal influenza in the production of inflammatory cytokines and chemokines.
Once influenza encounters the cell there are pattern recognition receptors (PRR) that are stimulated to initiate an antiviral response leading to the production of interferon and cytokines and chemokines. The three main pathways that are triggered leading to an interferon response are the toll like receptor pathway (TLR), retinoic acid inducible gene I (RIG-I) like receptors (RLR), and nucleotide oligomerization domain (NOD)-like receptor pathway pyrin domain containing 3 (NLRP3) (reviewed in van de Sandt [54]).
TLPR 2 and 4 are present on the cell surface and recognize viral surface glycoproteins, while TLR 3 and 7 are present intracellularly and recognized double stranded viral RNA (dsRNA) and single stranded RNA viral RNA9(ssRNA) respectively. Shinya and colleagues previously demonstrated that pre-stimulation of TLR2 and 4 had a protective effect in mice when challenged with HPAI and that this protective effect was also seen after pre-treatment with lipopolysaccharide (LPS) which is a ligand for TLR4 [55]. This TLR4 binding resulted in the activation of 2 independent pathways - one involving MyD88 and the other TRIF. This pathway also required 2 surface molecules MDM2 and CD14, and additional studies demonstrated that cultures lacking either MD2 or CD14 had no change in H5N1 replication after LPS stimulation [56]. Interestingly, while LPS alone did not affect the expression of interferon stimulation genes. It did lead to an upregulation of the expression of other antiviral molecules, and TLR3 was found to be upregulated in this TLR4-TRIF pathway, thus demonstrating that the suppression of H5N1 infection was aided by the synergistic upregulation of TLR3. This finding is of interest because TLR3 was previously thought to recognize only dsRNA [57].
TLR7, as mentioned before, is an intracellular receptor that recognizes ssRNA but only after the complex has been degraded in the endosome. Though it plays an important role in the innate defence mechanism, it is also involved in the antibody isotype switching and Katz and colleagues showed that TLR signaling was involved in the recruitment of myeloid-derived suppressor cells (MDSC) that play a role in the formation of the humoral immune response, directing the acute adaptive response towards Th2 following influenza virus infection [58]. Thus TLR7 appears to play a dual role - the recognition of vRNA in early infection and then in terms of immunization TLR7 is activated in the MHC class II endocytic pathway. As a backup to the late endosomal compartment there are 2 other pathways that recognize vRNA in the cytosolic component - RIG-I and NLR.
There are a number of good review articles on this pathway available [59-61]. These pathways exist in order to distinguish host "self" RNA from invading pathogenic RNA (i.e "non-self"). The intracellular detection of this RNA was ascribed to two cytoplasmic helicases called RIG-I and melanoma differentiation-associated gene 5 product (MDA5). RIG-I has 2 consecutive caspase activation and recruitment domains (CARD), a central helicase domain and a C-terminal domain (Fig. 3). Knockout studies showed that RIG-I and MDA5 were able to discriminate between different strains of virus - RIG-I for the ortho, paramyxo and rhabdoviruses and MDA5 for the picornaviruses [62].
It should be noted that there is a third member of the RLR pathway, LGP2 which lacks the CARD interaction motif. This is thought to act as a positive regulator making the viral RNA complexes more accessible to RIG-I. The recognition of viral RNA by RIG-I is enhanced when there is a 5'-triphosphate (PPP) present on the RNA, acting as a 'hook', as removal of this PPP reduced interferon induction [63, 64]. Once the triphosphate terminated vRNA is recognized by RIG-I, the complex undergoes a conformation change in which the CARDs are able to be ubiquitinated by TRIM25 which adds a Lys-63 linked ubiquitin chain to RIG-I. The Lys172 in the second CARD of RIG-I appears crucial for effective function [65] . Unlike many other results of ubiqutination, the change in RIG-I does not lead to degradation in the proteasome but actually is a critical step for subsequent interaction of RIG-I with MAVS though the exact mechanism how this is facilitated is not clearly understood [61].
The TRIM proteins are called such because they have a TRI-partite Motif and even though there have been over 70 genes encoding TRIM described, only a small number have been well characterized [61]. Since it was demonstrated that genes encoding TRIM were upregulated by type-1 interferons their role has been further investigated [66]. As mentioned above TRIM25 ubiquitinates and activates RIG-1, TRIM30α inhibits NF1κβ, TRIM21 stabilizes IRF3 and TRIM27 inhibits NF1κβ, IRF3 and IRF7, thus affecting the downstream activation pathway in viral infection. Once CARDs are expressed and ubiquitnated they are able to interact with the CARDs of the adaptor protein Mitochondrial AntiViral Signaling (MAVS) on mitochondria leading to activation of the IKKα-IKKβ-IKKγ complex and the downstream NFκβ, IRF3 and IRF7 complexes followed by transcription of type 1 interferons.
The end result of activation of the TLR and RIG-I pathway is the increased transcription of cytokines, chemokines and interferons that recruit neutrophils, activate macrophages and lead to maturation of dendritic cells. In influenza infection the type 1 interferons (IFN-α and IFN-β) are the major cytokines that limit viral replication with TNFα, IL-1β and IL-6 recruiting immune cells to the sites of infection and producing inflammation. The type-1 interferons act on INF-α/β receptors present on the same cell and on adjacent cells to activate the antiviral signaling cascade, including the Tyrosine Kinase 2 (TK2) and Janus Kinase 1 (JAK1), phosphorylation of STAT1/2 that indues transcription of hundreds of genes containing interferon stimulating response elements (IRSE)(reviewed by van de Sandt [54]). These IFN stimulated genes encode other TRIM proteins plus other antiviral proteins such as MxA, viperin, tetherin and OAS. One protein in particular PKR is involved in dsDNA binding and functions as an antiviral by blocking general translation.
Endothelial cells in the past were considered as collateral damage when the process of DAD was initiated as a loss of the endothelial integrity with subsequent microvascular leakage was a key feature in bacterial sepsis and in viral infection such as H5N1 [67]. In the 2009 pandemic autopsy studies demonstrated viral antigen in endothelial cells and if the same can be demonstrated in H5N1 infection this may explain the extrapulmonary dissemination of virus in these infections [8]. It has also been shown that polarized endothelial cells can be infected in vitro [68]. Intriguingly recent publications have shown that endothelial cells may actually be involved in modulating the innate immune response via sphingosine-1-phosphate signaling. This is a metabolite of sphingolipid and a ligand for G-protein coupled receptors SIP1-5 [69]. The role of these receptor ligands was shown by Marsolais and colleagues who used SIP ligands and found that they reduced the cytokine response using a mouse adapted influenza virus [70]. Further work demonstrated a level of SIP1 expression on lymphatic and vascular endothelial cells and that treatment with an agonist of SIP1 was able to reduce the production of chemokines CCL2, CCL5 and CD11b+ cells and decrease IFNγ secretion but did not affect IFNα, CCL2, IL6 or TNFα, thus showing that endothelial cells are able to actually modulate the inflammatory response [71]. It is possible that these SIP analogs may have a use in further therapies in dampening the excessive innate immune response [72].
The preceding discussion indicates that the human respiratory tract has developed a complex network of mechanism to ensure that it is protected from the effects of viral infection. The fact that severe influenza can occur in the lower respiratory tract shows that the virus has evolved mechanisms to circumvent this protective mechanism. These can broadly be divided into the surface proteins (HA and HA) that are involved in the extracellular phase of exposure (from the environment to the surface of the cell) and then once the virus is in the intracellular environment, the internal genes take over to enhance replication of progeny virions. With regard to the extacellular compartment the role of NA in cleaving the sialylated mucin and glycosylation of the HA has already been discussed.
Within the cell it has been found for a long time that influenza has evolved a strategy to evade the powerful IFN defence mechanism [73] but in 1998 Garcia-Sastre and colleagues showed that the production of recombinant influenza lacking a gene called non-structural protein (NS1) was able to suppress this IFN pathway [74]. NS1 employs this strategy at many levels, including targeting TRIM25. It will be remembered that TRIM25 is required to ubiqutinate and activate RIG-I leading to subsequent IFN production. For TRIM25 to perform this function it needs to be oligomerized and NS1 inhibition of this abolishes this activity. All influenza strains have this interaction with TRIM25, possibly because of the conserved Glu96 and Glu97 but the D92E change in H5N1 may affect function [75]. NS1 also functions to impair the interferon induced protein PKR which it will be recalled can shut down translation by sustained phosphorylation of the translation factor elF2α thus inhibiting virus propagation [76].
Finally a distinctive feature of the 1918 H1N1 NS1 and of many avian strains (including H5N1) is a C-terminal Glu-Ser-Glu-Val (ESEV) binding motif for a PDZ domain that mediates protein interaction. This finding was identified by large-scale sequence analysis of viruses isolated from different bird and mammalian species [77]. The ESEV domain is mainly present in avian influenza viruses while human viruses have a conserved RSKV domain. Jackson and colleagues in 2008 used reverse genetics to insert these different motifs into the mouse adapted A/WSN/33 and found that the RSKV had a better survival and lower viral load than the ESEV motif [78]. Later studies by Soubries and colleagues used LPAI H7N1 with a C-terminal RSKV and different species of cells and animals (ducks and mice), and found the changes in replication between ESEV and RSKV were species specific. In particular ESEV induced higher levels of IFN than RSKV in mouse cells but this ESKV was less sensitive to type 1 interferon pre-treatment [79]. The same groups also recently studied H7N1 HPAI with a deletion of 6 amino acids R225VESEV230 that was present in recent strains affecting poultry and found that the C-terminal ESEV domain did not increase virulence as deletion of the six amino acids had no impact on viral replication or immune response [80]. It should be noted that the 2008 H1N1 virus also has a stop codon at 220, thus suggesting that it behaves similar to H7N1 with the six amino acid deletion.
Finally, since there is evidence that severe influenza caused by certain influenza viruses such as H5N1 trigger an exaggerated innate immune response, are there therapeutic options in addition to antiviral therapy that may be utilized to "buy time" before the acquired immune response takes over? The use of immunmodulatory agents such as steroids has been discouraged by the WHO. Statin therapy has been advocated by a number of individuals and reviewed in a number of publications, with claimed evidence that there was a 66% reduction in mortality (reviewed by Fedson [81]) but no effect on viral clearance in infected mice [82]. One research group has used palmidronate to stimulate Vγ9Vδ2 T-cells to kill infected cells [83].An NF-κβ inhibitor SC75741 was shown to block H5N1 virus propagation in vitro and vivo and consequent overproduction of cytokines and chemokines in the lungs of mice after infection with H5N1 (Reiling SJ, unpublished observations). Evidence for a role for IFN in anti-influenza virus response has been shown through the use of IFNα/β receptor deficient mice [84].
IFNα has been shown as an effective control of hepatitis B and C in humans, particularly active HCV, in which IFNα has cured 98% of affected individuals. During the 2003 SARS outbreak in Toronto, IFN alfacon-1, a novel synthetic consensus IFN, showed clinical benefit in patients treated with steroids and IFN compared with those treated with steroids alone [85]. In guinea pigs and ferrets infected with H5N1 and seasonal influenza respectively, IFN treatment resulted in reduced viral titres and improved pulmonary pathology [86, 87]. There were promising results of intranasal administration of IFNα in preventing influenza infection in the 1980's but side-effects prevented the widespread adoption of this route of administration [88, 89]. Treatment with alfacon-1 of human ex vivo tissues infected with H1N1pdm and H5N1 showed it was able to reduce infection but IFN treatment post-exposure produced a less dramatic effect [90]. As this latter scenario is the one most likely to be encountered in clinical practice this implies that IFN use in the clinical setting may have only limited benefit.
In the event of a new pandemic with a severity and transmission similar to the 1918 H1N1 outbreak there will be limited availability of antiviral agents. A comprehensive understanding of pathways and mechanisms involved in the innate immune antiviral response is thus necessary so that additional agents may be used to control viral replication and spread until adequate vaccination is available.
Figures and Tables
Figure 1
The first barrier to infection - mucin.
Mucicarmine stained section of the normal human bronchus showing goblet cells and submucous glands containing purple staining mucin. Avian influenza viruses preferentially bind to goblet cells while human viruses bind to ciliated cells. Inset shows the interaction of viral haamagglutinin with the sialic acid terminated glycoprotein present on the surface of cells.
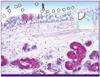
Figure 2
The second barrier to infection - host defence lectins.
Section of the normal human bronchiole and alveoli with red colour indicating the presence of 2-6 terminated sialic acid. Insets show surfactant protein A and D (SP-A and SP-D) bind to influenza viruses in two ways. The SP-A is sialylated and so acts as a decoy receptor. SP-D recognizes the galactose or mannose present on the surface of the influenza viruses. Macrophages also recognize the mannose present on viruses.
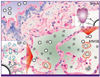
Figure 3
Intracellular activation of the RIG-I pathway.
Electron micrograph of human ex vivo tissues infected with avian influenza virus showing egress of viral particles from type 2 pneumocytes. The viral RNA binds to RIG-I and is then ubiquitinated by TRIM25 allowing binding to MAVS followed by activation of TBK-1, IKK pathways and transcription of interferon. The release of interferon leads to increased cytokine production, recruitment of neutrophils and activation of macrophages. The interferon will act as an autocrine and paracrine function leading to more cytokine and antiviral proteins.

Acknowledgments
Funding from Area of Excellence Scheme of the University Grants Committee (grant AoE/M-12/96).
References
1. Abrahams A, Hallows N, French H. A further investigation into influenza pneumococcal and influenza streptococcal septicaemia: epidemic influenza pneumonia of highly fatal type and its relation to purulent bronchitis. Lancet. 1919. 1:1–9.
2. Taubenberger JK. The origin and virulence of the 1918 "Spanish" influenza virus. Proc Am Philos Soc. 2006. 150:86–112.
3. Reid AH, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 "Spanish" influenza virus hemagglutinin gene. Proc Natl Acad Sci U S A. 1999. 96:1651–1656.

4. Tscherne DM, García-Sastre A. An enzymatic assay for detection of viral entry. Curr Protoc Cell Biol. 2011. Chapter 26:Unit 26.12.

5. Walsh JJ, Dietlein LF, Low FN, Burch GE, Mogabgab WJ. Bronchotracheal response in human influenza. Type A, Asian strain, as studied by light and electron microscopic examination of bronchoscopic biopsies. Arch Intern Med. 1961. 108:376–388.
6. Hers JF, Masurel N, Mulder J. Bacteriology and histopathology of the respiratory tract and lungs in fatal Asian influenza. Lancet. 1958. 2:1141–1143.

7. Guarner J, Paddock CD, Shieh WJ, Packard MM, Patel M, Montague JL, Uyeki TM, Bhat N, Balish A, Lindstrom S, Klimov A, Zaki SR. Histopathologic and immunohistochemical features of fatal influenza virus infection in children during the 2003-2004 season. Clin Infect Dis. 2006. 43:132–140.

8. Shieh WJ, Blau DM, Denison AM, Deleon-Carnes M, Adem P, Bhatnagar J, Sumner J, Liu L, Patel M, Batten B, Greer P, Jones T, Smith C, Bartlett J, Montague J, White E, Rollin D, Gao R, Seales C, Jost H, Metcalfe M, Goldsmith CS, Humphrey C, Schmitz A, Drew C, Paddock C, Uyeki TM, Zaki SR. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol. 2010. 177:166–175.
9. Uiprasertkul M, Puthavathana P, Sangsiriwut K, Pooruk P, Srisook K, Peiris M, Nicholls JM, Chokephaibulkit K, Vanprapar N, Auewarakul P. Influenza A H5N1 replication sites in humans. Emerg Infect Dis. 2005. 11:1036–1041.

10. Gu J, Xie Z, Gao Z, Liu J, Korteweg C, Ye J, Lau LT, Lu J, Gao Z, Zhang B, McNutt MA, Lu M, Anderson VM, Gong E, Yu AC, Lipkin WI. H5N1 infection of the respiratory tract and beyond: a molecular pathology study. Lancet. 2007. 370:1137–1145.

11. Liem NT, Nakajima N, Phat le P, Sato Y, Thach HN, Hung PV, San LT, Katano H, Kumasaka T, Oka T, Kawachi S, Matsushita T, Sata T, Kudo K, Suzuki K. H5N1-infected cells in lung with diffuse alveolar damage in exudative phase from a fatal case in Vietnam. Jpn J Infect Dis. 2008. 61:157–160.
12. Writing Committee of the Second World Health Organization Consultation on Clinical Aspects of Human Infection with Avian Influenza A (H5N1) Virus. Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, Naghdaliyev A, Peiris JS, Shindo N, Soeroso S, Uyeki TM. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008. 358:261–273.

13. Suptawiwat O, Tantilipikorn P, Boonarkart C, Lumyongsatien J, Uiprasertkul M, Puthavathana P, Auewarakul P. Enhanced susceptibility of nasal polyp tissues to avian and human influenza viruses. PLoS One. 2010. 5:e12973.

14. Zhang Z, Zhang J, Huang K, Li KS, Yuen KY, Guan Y, Chen H, Ng WF. Systemic infection of avian influenza A virus H5N1 subtype in humans. Hum Pathol. 2009. 40:735–739.

15. Nicholls JM, Butany J, Poon LL, Chan KH, Beh SL, Poutanen S, Peiris JS, Wong M. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. 2006. 3:e27.

16. Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, Liu H, Sun Y, Pasparakis M, Kopf M, Mech C, Bavari S, Peiris JS, Slutsky AS, Akira S, Hultqvist M, Holmdahl R, Nicholls J, Jiang C, Binder CJ, Penninger JM. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008. 133:235–249.

17. Hirst GK. Adsorption of influenza hemagglutinins and virus by red blood Cells. J Exp Med. 1942. 76:195–209.

19. Varki NM, Varki A. Diversity in cell surface sialic acid presentations: implications for biology and disease. Lab Invest. 2007. 87:851–857.

21. Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983. 127:361–373.

22. Baum LG, Paulson JC. Sialyloligosaccharides of the respiratory epithelium in the selection of human influenza virus receptor specificity. Acta Histochem Suppl. 1990. 40:35–38.
23. Scholtissek C, Burger H, Kistner O, Shortridge KF. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985. 147:287–294.

24. Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998. 72:7626–7631.

25. Trebbien R, Larsen LE, Viuff BM. Distribution of sialic acid receptors and influenza A virus of avian and swine origin in experimentally infected pigs. Virol J. 2011. 8:434.

26. Nelli RK, Kuchipudi SV, White GA, Perez BB, Dunham SP, Chang KC. Comparative distribution of human and avian type sialic acid influenza receptors in the pig. BMC Vet Res. 2010. 6:4.

27. Van Poucke SG, Nicholls JM, Nauwynck HJ, Van Reeth K. Replication of avian, human and swine influenza viruses in porcine respiratory explants and association with sialic acid distribution. Virol J. 2010. 7:38.

29. Williams OW, Sharafkhaneh A, Kim V, Dickey BF, Evans CM. Airway mucus: From production to secretion. Am J Respir Cell Mol Biol. 2006. 34:527–536.
30. Wheeler AH, Nungester WJ. Effect of mucin on influenza virus infection in hamsters. Science. 1942. 96:92–93.

31. Lo-Guidice JM, Merten MD, Lamblin G, Porchet N, Houvenaghel MC, Figarella C, Roussel P, Perini JM. Mucins secreted by a transformed cell line derived from human tracheal gland cells. Biochem J. 1997. 326:431–437.

32. Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993. 29:155–165.

33. Nicholls JM, Bourne AJ, Chen H, Guan Y, Peiris JS. Sialic acid receptor detection in the human respiratory tract: evidence for widespread distribution of potential binding sites for human and avian influenza viruses. Respir Res. 2007. 8:73.

34. Ng WC, Tate MD, Brooks AG, Reading PC. Soluble host defense lectins in innate immunity to influenza virus. J Biomed Biotechnol. 2012. 2012:732191.

35. Basak S, Tomana M, Compans RW. Sialic acid is incorporated into influenza hemagglutinin glycoproteins in the absence of viral neuraminidase. Virus Res. 1985. 2:61–68.

36. Sun S, Wang Q, Zhao F, Chen W, Li Z. Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS One. 2011. 6:e22844.

37. Reading PC, Tate MD, Pickett DL, Brooks AG. Glycosylation as a target for recognition of influenza viruses by the innate immune system. Adv Exp Med Biol. 2007. 598:279–292.

38. Pritchett TJ, Paulson JC. Basis for the potent inhibition of influenza virus infection by equine and guinea pig alpha 2-macroglobulin. J Biol Chem. 1989. 264:9850–9858.

39. Kingma PS, Zhang L, Ikegami M, Hartshorn K, McCormack FX, Whitsett JA. Correction of pulmonary abnormalities in Sftpd-/- mice requires the collagenous domain of surfactant protein D. J Biol Chem. 2006. 281:24496–24505.

40. Reading PC, Allison J, Crouch EC, Anders EM. Increased susceptibility of diabetic mice to influenza virus infection: compromise of collectin-mediated host defense of the lung by glucose? J Virol. 1998. 72:6884–6887.

41. Ward KA, Spokes PJ, McAnulty JM. Case-control study of risk factors for hospitalization caused by pandemic (H1N1) 2009. Emerg Infect Dis. 2011. 17:1409–1416.

42. Peiris JS, Yu WC, Leung CW, Cheung CY, Ng WF, Nicholls JM, Ng TK, Chan KH, Lai ST, Lim WL, Yuen KY, Guan Y. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004. 363:617–619.

43. Nicholls JM, Chan MC, Chan WY, Wong HK, Cheung CY, Kwong DL, Wong MP, Chui WH, Poon LL, Tsao SW, Guan Y, Peiris JS. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med. 2007. 13:147–149.

44. Yu WC, Chan RW, Wang J, Travanty EA, Nicholls JM, Peiris JS, Mason RJ, Chan MC. Viral replication and innate host responses in primary human alveolar epithelial cells and alveolar macrophages infected with influenza H5N1 and H1N1 viruses. J Virol. 2011. 85:6844–6855.

45. van Riel D, Leijten LM, van der Eerden M, Hoogsteden HC, Boven LA, Lambrecht BN, Osterhaus AD, Kuiken T. Highly pathogenic avian influenza virus H5N1 infects alveolar macrophages without virus production or excessive TNF-alpha induction. PLoS Pathog. 2011. 7:e1002099.

46. Upham JP, Pickett D, Irimura T, Anders EM, Reading PC. Macrophage receptors for influenza A virus: role of the macrophage galactose-type lectin and mannose receptor in viral entry. J Virol. 2010. 84:3730–3737.

47. Hwang I, Scott JM, Kakarla T, Duriancik DM, Choi S, Cho C, Lee T, Park H, French AR, Beli E, Gardner E, Kim S. Activation mechanisms of natural killer cells during influenza virus infection. PLoS One. 2012. 7:e51858.

48. Crouch E, Hartshorn K, Ofek I. Collectins and pulmonary innate immunity. Immunol Rev. 2000. 173:52–65.

49. Hillaire ML, van Eijk M, Nieuwkoop NJ, Vogelzang-van Trierum SE, Fouchier RA, Osterhaus AD, Haagsman HP, Rimmelzwaan GF. The number and position of N-linked glycosylation sites in the hemagglutinin determine differential recognition of seasonal and 2009 pandemic H1N1 influenza virus by porcine surfactant protein D. Virus Res. 2012. 169:301–305.

50. van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RA, Osterhaus AD, Kuiken T. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol. 2007. 171:1215–1223.

51. Weinheimer VK, Becher A, Tönnies M, Holland G, Knepper J, Bauer TT, Schneider P, Neudecker J, Rückert JC, Szymanski K, Temmesfeld-Wollbrueck B, Gruber AD, Bannert N, Suttorp N, Hippenstiel S, Wolff T, Hocke AC. Influenza A viruses target type II pneumocytes in the human lung. J Infect Dis. 2012. 206:1685–1694.

52. Hampton T. Virulence of 1918 influenza virus linked to inflammatory innate immune response. JAMA. 2007. 297:580.

53. Chan MC, Cheung CY, Chui WH, Tsao SW, Nicholls JM, Chan YO, Chan RW, Long HT, Poon LL, Guan Y, Peiris JS. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir Res. 2005. 6:135.

54. van de Sandt CE, Kreijtz JH, Rimmelzwaan GF. Evasion of influenza a viruses from innate and adaptive immune responses. Viruses. 2012. 4:1438–1476.

55. Shinya K, Okamura T, Sueta S, Kasai N, Tanaka M, Ginting TE, Makino A, Eisfeld AJ, Kawaoka Y. Toll-like receptor pre-stimulation protects mice against lethal infection with highly pathogenic influenza viruses. Virol J. 2011. 8:97.

56. Shinya K, Ito M, Makino A, Tanaka M, Miyake K, Eisfeld AJ, Kawaoka Y. The TLR4-TRIF pathway protects against H5N1 influenza virus infection. J Virol. 2011. 86:19–24.

57. Guillot L, Le Goffic R, Bloch S, Escriou N, Akira S, Chignard M, Si-Tahar M. Involvement of toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J Biol Chem. 2005. 280:5571–5580.

58. Jeisy-Scott V, Kim JH, Davis WG, Cao W, Katz JM, Sambhara S. TLR7 recognition is dispensable for influenza virus A infection but important for the induction of hemagglutinin-specific antibodies in response to the 2009 pandemic split vaccine in mice. J Virol. 2012. 86:10988–10998.

59. Bowzard JB, Davis WG, Jeisy-Scott V, Ranjan P, Gangappa S, Fujita T, Sambhara S. PAMPer and tRIGer: ligand-induced activation of RIG-I. Trends Biochem Sci. 2011. 36:314–319.

60. Wolff T, Zielecki F, Abt M, Voss D, Semmler I, Matthaei M. Sabotage of antiviral signaling and effectors by influenza viruses. Biol Chem. 2008. 389:1299–1305.

62. Ramos I, Fernandez-Sesma A. Cell receptors for influenza a viruses and the innate immune response. Front Microbiol. 2012. 3:117.

63. Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006. 314:994–997.

64. Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006. 314:997–1001.

65. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007. 446:916–920.

66. Ozato K, Shin DM, Chang TH, Morse HC 3rd. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008. 8:849–860.

67. Steinberg BE, Goldenberg NM, Lee WL. Do viral infections mimic bacterial sepsis? The role of microvascular permeability: A review of mechanisms and methods. Antiviral Res. 2012. 93:2–15.

68. Chan MC, Chan RW, Yu WC, Ho CC, Chui WH, Lo CK, Yuen KM, Guan YI, Nicholls JM, Peiris JS. Influenza H5N1 virus infection of polarized human alveolar epithelial cells and lung microvascular endothelial cells. Respir Res. 2009. 10:102.

70. Marsolais D, Hahm B, Walsh KB, Edelmann KH, McGavern D, Hatta Y, Kawaoka Y, Rosen H, Oldstone MB. A critical role for the sphingosine analog AAL-R in dampening the cytokine response during influenza virus infection. Proc Natl Acad Sci U S A. 2009. 106:1560–1565.

71. Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB, Rosen H. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011. 146:980–991.

72. Walsh KB, Teijaro JR, Rosen H, Oldstone MB. Quelling the storm: utilization of sphingosine-1-phosphate receptor signaling to ameliorate influenza virus-induced cytokine storm. Immunol Res. 2011. 51:15–25.

73. Kochs G, García-Sastre A, Martínez-Sobrido L. Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol. 2007. 81:7011–7021.

74. García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998. 252:324–330.

75. Li M, Wang B. Homology modeling and examination of the effect of the D92E mutation on the H5N1 nonstructural protein NS1 effector domain. J Mol Model. 2007. 13:1237–1244.

76. García MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007. 89:799–811.

77. Chen GW, Chang SC, Mok CK, Lo YL, Kung YN, Huang JH, Shih YH, Wang JY, Chiang C, Chen CJ, Shih SR. Genomic signatures of human versus avian influenza A viruses. Emerg Infect Dis. 2006. 12:1353–1360.

78. Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008. 105:4381–4386.

79. Soubies SM, Volmer C, Croville G, Loupias J, Peralta B, Costes P, Lacroux C, Guérin JL, Volmer R. Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J Virol. 2010. 84:6733–6747.

80. Soubies SM, Hoffmann TW, Croville G, Larcher T, Ledevin M, Soubieux D, Quéré P, Guérin JL, Marc D, Volmer R. Deletion of the C-terminal ESEV domain of NS1 does not affect the replication of a low-pathogenic avian influenza virus H7N1 in ducks and chickens. J Gen Virol. 2013. 94:50–58.

81. Fedson DS. Confronting the next influenza pandemic with anti-inflammatory and immunomodulatory agents: why they are needed and how they might work. Influenza Other Respi Viruses. 2009. 3:129–142.

82. Radigan KA, Urich D, Misharin AV, Chiarella SE, Soberanes S, Gonzalez A, Perlman H, Wunderink RG, Budinger GR, Mutlu GM. The effect of rosuvastatin in a murine model of influenza A infection. PLoS One. 2012. 7:e35788.

83. Tu W, Zheng J, Liu Y, Sia SF, Liu M, Qin G, Ng IH, Xiang Z, Lam KT, Peiris JS, Lau YL. The aminobisphosphonate pamidronate controls influenza pathogenesis by expanding a gammadelta T cell population in humanized mice. J Exp Med. 2011. 208:1511–1522.

84. Szretter KJ, Gangappa S, Belser JA, Zeng H, Chen H, Matsuoka Y, Sambhara S, Swayne DE, Tumpey TM, Katz JM. Early control of H5N1 influenza virus replication by the type I interferon response in mice. J Virol. 2009. 83:5825–5834.

85. Loutfy MR, Blatt LM, Siminovitch KA, Ward S, Wolff B, Lho H, Pham DH, Deif H, LaMere EA, Chang M, Kain KC, Farcas GA, Ferguson P, Latchford M, Levy G, Dennis JW, Lai EK, Fish EN. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA. 2003. 290:3222–3228.

86. Van Hoeven N, Belser JA, Szretter KJ, Zeng H, Staeheli P, Swayne DE, Katz JM, Tumpey TM. Pathogenesis of 1918 pandemic and H5N1 influenza virus infections in a guinea pig model: antiviral potential of exogenous alpha interferon to reduce virus shedding. J Virol. 2009. 83:2851–2861.

87. Kugel D, Kochs G, Obojes K, Roth J, Kobinger GP, Kobasa D, Haller O, Staeheli P, von Messling V. Intranasal administration of alpha interferon reduces seasonal influenza A virus morbidity in ferrets. J Virol. 2009. 83:3843–3851.

88. Finter NB, Chapman S, Dowd P, Johnston JM, Manna V, Sarantis N, Sheron N, Scott G, Phua S, Tatum PB. The use of interferon-alpha in virus infections. Drugs. 1991. 42:749–765.




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download