Abstract
Background
We evaluated the ability of infrequent restriction site-polymerase chain reaction (IRS-PCR) to perform molecular epidemiologic analysis of Community-Onset Extended Spectrum Beta-Lactamase (ESBL) producing Escherichia coli, and also assessed the use of PFGE as an alternative method.
Materials and Methods
IRS-PCR assay was performed using combinations of adaptors for XbaI and HhaI restriction sites on clinical isolates of E. coli (n=51). We compared the discriminatory power, quality and efficiency of IRS-PCR to PFGE.
Figures and Tables
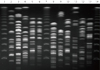 | Figure 1Representative Pulsed-Field Gel Electrophoresis (PFGE) images of ESBL Escherichia coli isolates. Lanes 3 to 6 and 8 to 11 were BL430, BL435, BL440, BL441, BL442, BL443, BL446, BL447, BL448, BL450, and BL451, respectively. Markers were in lanes 1, 7 and 14 (50-1,000kb ladders). |
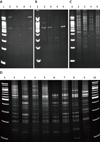 | Figure 2Electrophoretic results of IRS-PCR amplification of restricted-ligated XbaI-HhaI fragments from ESBL strains. (A) Lane 1 was 100-bp ladder. Lanes 2 through 5 were isolates of BL99, BL101, BL102 and BL105, respectively. Clinically isolated strains used adaptors (AH1 and AX2) and primer PX-G. Tm; 20.6℃, 14.3℃. (B) Lane 1 was 100-bp ladder. Lanes 2 through 5 were isolates of BL99, BL101, BL102 and BL105, respectively. Clinically isolated strains used adaptors (AH1 and AH2) and primer PX-G. Tm; 32.6℃, 27.3℃. (C) Lane 1 was 100-bp ladder. Lanes 2 through 5 were isolates of BL99, BL101, BL102, and BL105, respectively. Clinically isolated strains used adaptors (AH1_new and AH2_new) and primer PX-G. Tm; 51.4℃, 37.4℃. (D) Representative IRS-PCR electrophoresis patterns of eight ESBL E. coli isolates. Lanes 2 through 9 were BL99, BL101, BL102, BL105, BL107, BL114, BL116 and BL117, respectively. Lanes 1 and 10 were 100bp ladders. The IRS-PCR annealing temperature was 58℃, and the primers were PX-G, AH2_new and AX2_new. |
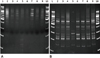 | Figure 3IRS-PCR electrophoretic patterns of ESBL E.coli produced by amplification of restricted-ligated XbaI-HhaI fragments. (A) Lane 1 and 10 were 100bp ladders. Lanes 2 through 9 were isolates BL99, BL101, BL102, BL105, BL107, BL114, BL116 and BL117, respectively. Clinically isolated strains used adaptors (AH2_new and AX2_new) and primer PX-G. Room temperature ligation performed at 20-25℃. (B) Lane 1 and 10 were 100bp ladders. Lanes 2 through 9 were isolates BL99, BL101, BL102, BL105, BL107, BL114, BL116 and BL117, respectively. Clinically isolated strains used adaptors (AH2_new and AX2_new) and primer PX-G. Room temperature ligation performed at -16℃. |
References
1. Kim YK, Pai H, Lee HJ, Park SE, Choi EH, Kim J, Kim JH, Kim EC. Bloodstream infections by extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in children: epidemiology and clinical outcome. Antimicrob Agents Chemother. 2002. 46:1481–1491.

2. Lee SH, Kim JY, Shin SH, An YJ, Choi YW, Jung YC, Jung HI, Sohn ES, Jeong SH, Lee KJ. Dissemination of SHV-12 and characterization of new AmpC-type beta-lactamase genes among clinical isolates of enterobacter species in Korea. J Clin Microbiol. 2003. 41:2477–2482.

3. Pai H, Lee HJ, Choi EH, Kim J, Jacoby GA. Evolution of TEM-related extended-spectrum beta-lactamases in Korea. Antimicrob Agents Chemother. 2001. 45:3651–3653.
4. Pai H, Lyu S, Lee JH, Kim J, Kwon Y, Kim JW, Choe KW. Survey of extended-spectrum beta-lactamases in clinical isolates of Escherichia coli and Klebsiella pneumoniae: prevalence of TEM-52 in Korea. J Clin Microbiol. 1999. 37:1758–1763.
5. Livermore DM, Canton R, Gniadkowski M, Nordmann P, Rossolini GM, Arlet G, Ayala J, Coque TM, Kern-Zdanowicz I, Luzzaro F, Poirel L, Woodford N. CTX-M: changing the face of ESBLs in Europe. J Antimicrob Chemother. 2007. 59:165–174.

6. Rossolini GM, D'Andrea MM, Mugnaioli C. The spread of CTX-M-type extended-spectrum beta-lactamases. Clin Microbiol Infect. 2008. 14:Suppl 1. 33–41.

7. Shin WS. Molecular epidemiologic typing Boonja yeokhakjeok hyungbyul. Korean J Infect Dis. 1995. 27:585–594.

8. Mazurek GH, Hartman S, Zhang Y, Brown BA, Hector JS, Murphy D, Wallace RJ Jr. Large DNA restriction fragment polymorphism in the Mycobacterium avium-M. intracellulare complex: a potential epidemiologic tool. J Clin Microbiol. 1993. 31:390–394.

9. Mazurek GH, Reddy V, Marston BJ, Haas WH, Crawford JT. DNA fingerprinting by infrequent-restriction-site amplification. J Clin Microbiol. 1996. 34:2386–2390.

10. Yoo JH. Appbcation of molecular epidemiologic typing to the control of nosocomial infection. Korean J Nosocomial Infect Control. 1997. 2:61–71.

11. Bisharat N, Agmon V, Finkelstein R, Raz R, Ben-Dror G, Lerner L, Soboh S, Colodner R, Cameron DN, Wykstra DL, Swerdlow DL, Farmer JJ 3rd. Israel Vibrio Study Group. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet. 1999. 354:1421–1424.
12. Abi-Said D, Anaissie E, Uzun O, Raad I, Pinzcowski H, Vartivarian S. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin Infect Dis. 1997. 24:1122–1128.
13. Zhang J, Hollis RJ, Pfaller MA. Variations in DNA subtype and antifungal susceptibility among clinical isolates of Candida tropicalis. Diagn Microbiol Infect Dis. 1997. 27:63–67.

14. Kim SI, Yoo JH, Cho YK, Lee DG, Wie SH, Choi JH, Kim YR, Shin WS, Kang MW. Comparison of pulsed-field gel electrophoresis, amplified fragment length polymorphism, and infrequent restriction site-polymerase chain reaction for molecular typing of Escherichia coil and Staphylococcus aureus strains. Korean J Infect Dis. 1999. 31:474–480.





 PDF
PDF ePub
ePub Citation
Citation Print
Print






 XML Download
XML Download