Abstract
Human gut microbial community is playing a critical role in human health and associated with different human disease. In parallel, probiotics, antibiotics, and antipyretic analgesics (AAs) were developed to improve human health or cure human diseases. We therefore examined how probiotics, antibiotics, and AAs influence to the gut microbiota. Three independent case/control studies were designed from the cross-sectional cohort data of 1,463 healthy Koreans. The composition of the gut microbiota in each case and control group was determined via 16S ribosomal RNA Illumina next-generation sequencing. The correlation between microbial taxa and the consumption of each drug was tested using zero-inflated Gaussian mixture models, with covariate adjustment of age, sex, and body mass index (BMI). Probiotics, antibiotics, and AAs consumption yielded the significant differences in the gut microbiota, represented the lower abundance of Megasphaera in probiotics, the higher abundance of Fusobacteria in antibiotics, and the higher abundance of Butyrivibrio and Verrucomicrobia in AAs, compared to each control group. The reduction of Erysipelotrichaceae family was common in three drugs consumption.
Go to : 
REFERENCES
1). Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009; 9:313–23.

2). Clemente JC, Ursell LK, Parfrey LW, Knight R. The Impact of the Gut Microbiota on Human Health: An Integrative View. Cell. 2012; 148:1258–70.

3). Ianiro G, Tilg H, Gasbarrini A. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. 2016; 65:1906–15.

4). Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010; 7:503–14.

5). Angelakis E, Merhej V, Raoult D. Related actions of probiotics and antibiotics on gut microbiota and weight modification. Lancet Infect Dis. 2013; 13:889–99.

6). Rogers MA, Aronoff DM. The influence of nonsteroidal anti-inflammatory drugs on the gut microbiome. Clin Microbiol Infect. 2016; 22:178. e1-9.

7). van Baarlen P, Wells JM, Kleerebezem M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 2013; 34:208–15.

8). Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014; 124:4212–8.

9). Morgun A, Dzutsev A, Dong X, Greer RL, Sexton DJ, Ravel J, et al. Uncovering effects of antibiotics on the host and microbiota using transkingdom gene networks. Gut. 2015; 64:1732–43.

10). Peterson CT, Sharma V, Elmén L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol. 2015; 179:363–77.

11). Hinz B, Brune K. Antipyretic Analgesics: Nonsteroidal Antiinflammatory Drugs, Selective COX-2 Inhibitors, Paracetamol and Pyrazolinones. Analgesia. C Stein, editor. Berlin, Heidelberg: Springer Berlin Heidelberg;2007. p. 65–93.

12). Syer SD, Wallace JL. Environmental and NSAID-Enteropathy: Dysbiosis as a Common Factor. Curr Gastroenterol Rep. 2014; 16:377.

13). Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014; 2:6.

14). Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013; 10:996–8.

15). Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013; 531:371–444.

16). Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013; 10:1200–2.

17). Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007; 1:56–66.

19). Joyce SA, Gahan CG. The gut microbiota and the metabolic health of the host. Curr Opin Gastroenterol. 2014; 30:120–7.

20). Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol. 2005; 71:8228–35.

21). Hansen TA. Acidaminobacter. Bergey's Manual of Systematics of Archaea and Bacteria:. John Wiley & Sons, Ltd;2015.

22). Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic Health: Fermentation and Short Chain Fatty Acids. J Clin Gastroenterol. 2006; 40:235–43.

23). den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013; 54:2325–40.

24). Di Rienzi SC, Sharon I, Wrighton KC, Koren O, Hug LA, Thomas BC, et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife. 2013; 2:e01102.

25). Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014; 588:4223–33.

26). Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016; 8:42.

27). Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012; 22:299–306.

28). Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012; 22:292–8.

29). McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium Is Associated with Colorectal Adenomas. PLoS ONE. 2013; 8:e53653.

30). Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014; 74:1311–8.
31). Million M, Maraninchi M, Henry M, Armougom F, Richet H, Carrieri P, et al. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int J Obes. 2012; 36:817–25.

32). Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, et al. Human Genetics Shape the Gut Microbiome. Cell. 2014; 159:789–99.

33). Allen SJ, Martinez EG, Gregorio GV, Dans LF. Cochrane Database of Systematic Reviews-Probiotics for treating acute infectious diarrhoea:. John Wiley & Sons, Ltd;2010.
34). Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012; 10:323–35.

35). Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol. 2014; 10:416–24.

36). Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016; 65:426–36.
37). Duffin R, O'Connor RA, Crittenden S, Forster T, Yu C, Zheng X, et al. Prostaglandin E2 constrains systemic inflammation through an innate lymphoid cell-IL-22 axis. Science. 2016; 351:1333–8.
Go to : 
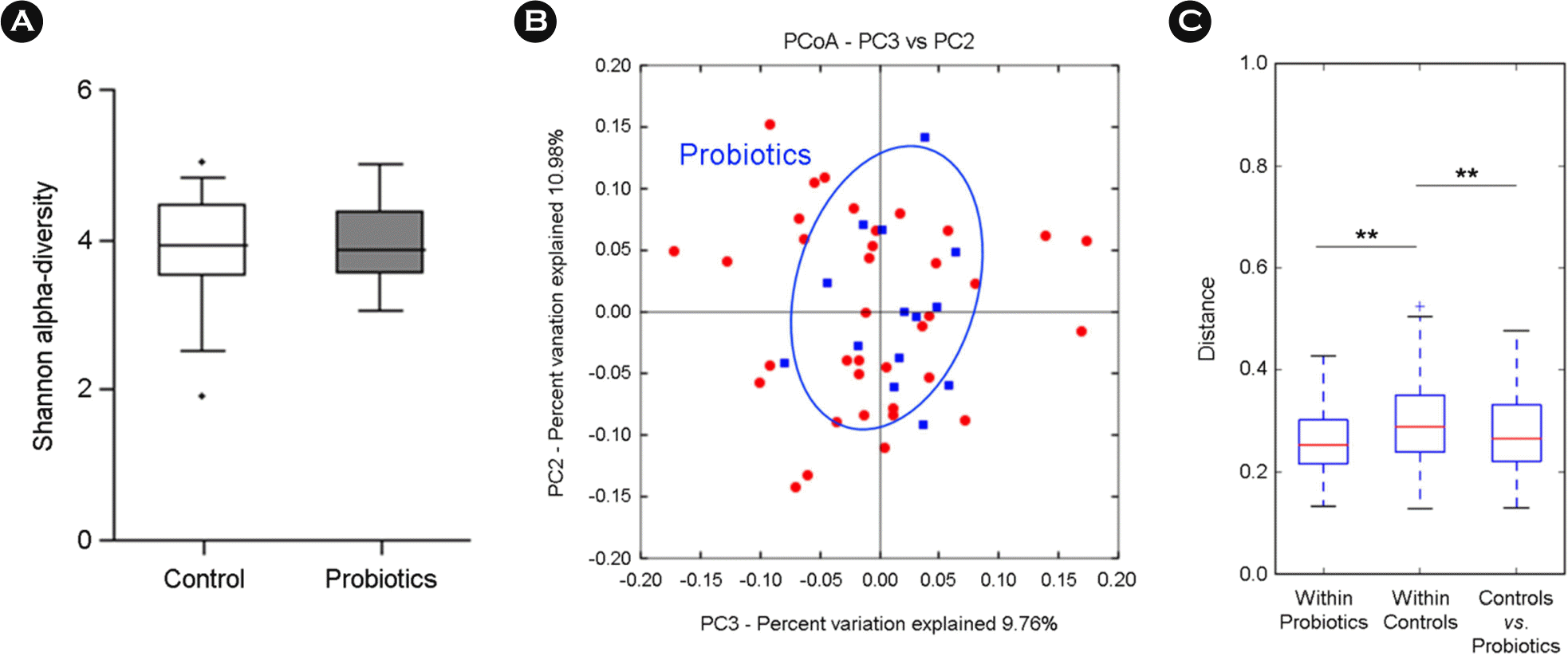 | Figure 1.Comparison of gut microbiota between 14 probiotics and 28 control groups. (A) Alpha-diversity (Shannon index), (B) beta-diversity of PCoA plots in probiotics group by weighted UniFrac, (C) weighted UniFrac distance box plot within and between groups (∗∗ p < 0.01). |
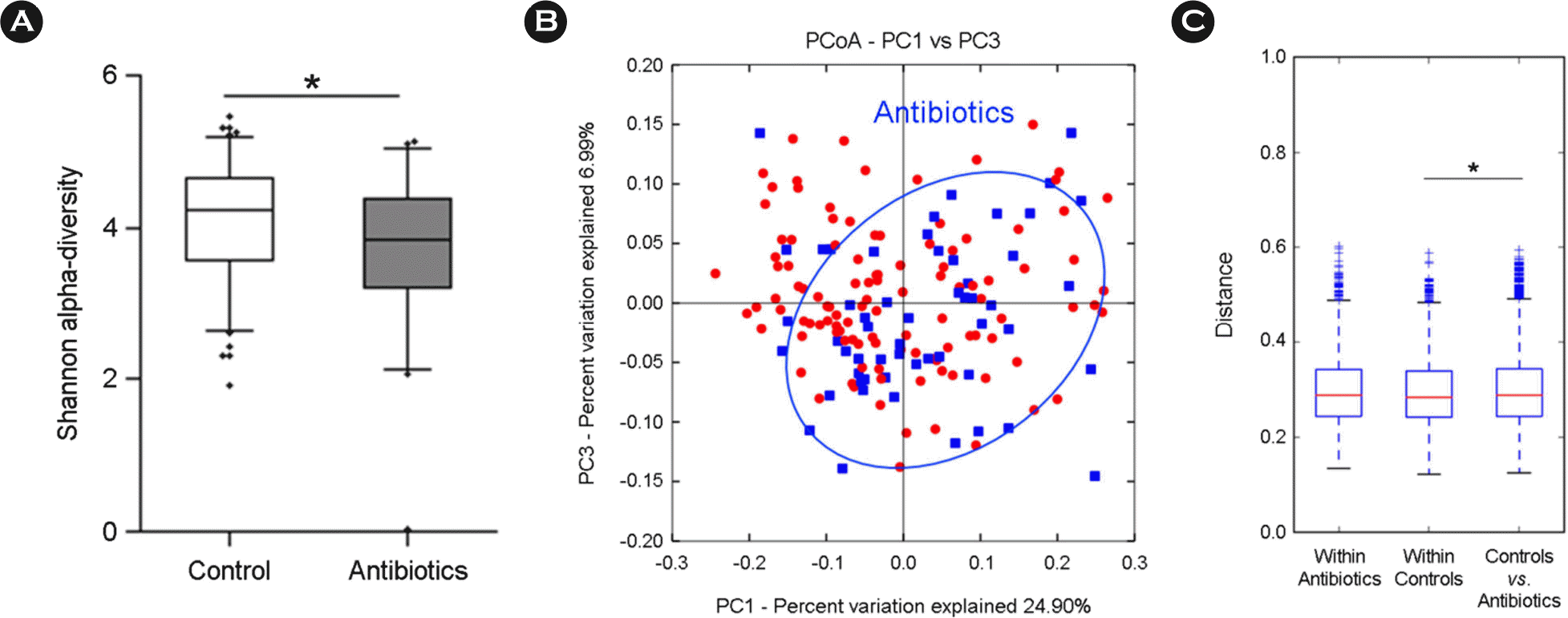 | Figure 2.Comparison of gut microbiota between 55 antibiotics and 110 control groups. (A) Alpha-diversity (Shannon index), (B) beta-diversity of PCoA plots in antibiotics group by weighted UniFrac, (C) weighted UniFrac distance box plot within and between groups (∗p < 0.05). |
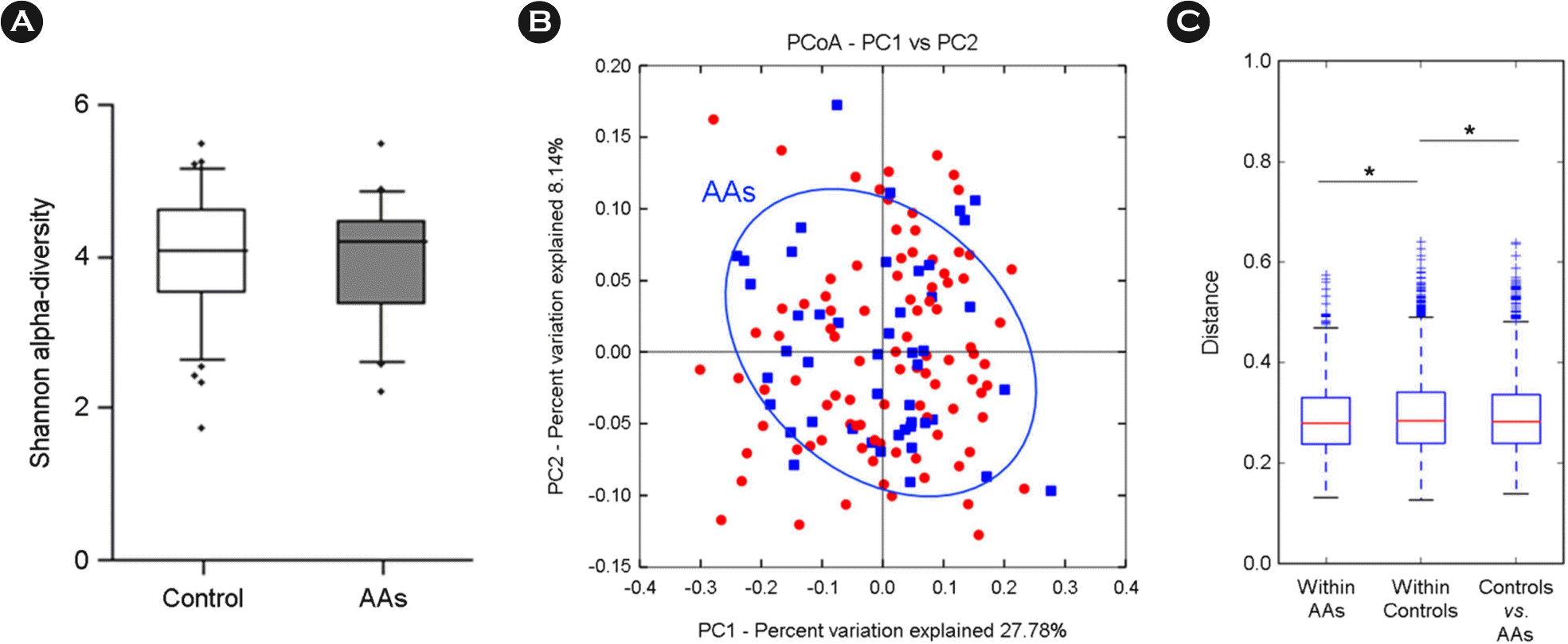 | Figure 3.Comparison of gut microbiota between 47 AAs and 94 control groups. (A) Alpha-diversity (Shannon index), (B) beta-diversity of PCoA plots in AAs group by weighted UniFrac, (C) weighted UniFrac distance box plot within and between groups (∗p < 0.05). |
Table 1.
Characteristics of probiotics study population
Table 2.
Characteristics of antibiotics study population
Table 3.
Characteristics of antipyretic analgesics (AAs) study population
Table 4.
Significant taxa profiles of gut microbiota assessed by 16S metagenomics sequencing related with probiotics consumption
| Phylum | Family | Genus | Coeff.a | p valueb |
|---|---|---|---|---|
| Cyanobacteria/Chloroplast | -2.37 | 0.017 | ||
| Cyanobacteria/Chloroplast | Streptophyta | -2.33 | 0.065 | |
| Firmicutes | Erysipelotrichaceae | Erysipelotrichaceae incertae sedis | -3.81 | 0.059 |
| Firmicutes | Veillonellaceae | Megasphaera | -6.53 | 0.003 |
Table 5.
Significant taxa profiles of gut microbiota assessed by 16S metagenomics sequencing related with antibiotics consumption
| Phylum | Family | Genus | Coeff.a | p valueb |
|---|---|---|---|---|
| Euryarchaeota (Archaea) | -2.97 | 6.04E-08 | ||
| Euryarchaeota (Archaea) | Methanobacteriaceae | -3.65 | 1.51E-10 | |
| Euryarchaeota (Archaea) | Methanobacteriaceae | Methanobrevibacter | -3.30 | 4.33E-09 |
| Bacterodetes | Porphyromonadaceae | Butyricimonas | -1.70 | 5.59E-04 |
| Firmicutes | Paenibacillaceae | -2.85 | 8.06E-12 | |
| Firmicutes | Paenibacillaceae | Paenibacillus | -2.85 | 1.11E-11 |
| Firmicutes | Christensenellaceae | -1.83 | 8.78E-06 | |
| Firmicutes | Christensenellaceae | Christensenella | -1.77 | 3.71E-05 |
| Firmicutes | Vallitalea | -1.59 | 0.01 | |
| Firmicutes | Enterococcaceae | -1.56 | 0.003 | |
| Firmicutes | Enterococcaceae | Enterococcus | -1.61 | 0.005 |
| Firmicutes | Acidaminococcaceae | Acidaminococcus | -1.75 | 0.025 |
| Firmicutes | Erysipelotrichaceae | Erysipelotrichaceae incertae sedis | -4.51 | 3.28E-17 |
| Firmicutes | Erysipelotrichaceae | Erysipelothrix | -4.07 | 1.17E-16 |
| Firmicutes | Lachnospiraceae | Butyrivibrio | -3.24 | 8.28E-08 |
| Firmicutes | Ruminococcaceae | Fastidiosipila | -1.55 | 1.97E-06 |
| Firmicutes | Ruminococcaceae | Anaerotruncus | -0.93 | 0.027 |
| Firmicutes | Veillonellaceae | Megasphaera | -2.50 | 9.54E-05 |
| Fusobacteria | 1.98 | 0.007 |
Table 6.
Significant taxa profiles of gut microbiota assessed by 16S metagenomics sequencing related with AAs consumption
| Phylum | Family | Genus | Coeff.a | p valueb |
|---|---|---|---|---|
| Euryarchaeota (Archaea) | 3.10 | 3.34E-05 | ||
| Verrucomicrobia | 1.57 | 0.015 | ||
| Firmicutes | Clostridiaceae | 1.77 | 0.026 | |
| Bacterodetes | Porphyromonadaceae | Butyricimonas | -2.41 | 2.54E-08 |
| Firmicutes | Lachnospiraceae | Butyrivibrio | 4.84 | 1.60E-06 |
| Firmicutes | Erysipelotrichaceae | Erysipelotrichaceae incertae sedis | -3.36 | 1.70E-06 |
| Proteobacteria | Desulfovibrionaceae | Desulfovibrio | 2.57 | 6.14E-06 |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download