Abstract
Urinary tract infections (UTI) are one of the most frequent infectious diseases. Uropathogenic Escherichia coli (UPEC) are among major pathogens causing UTI. A variety of virulence genes are mainly responsible for the severity of these emerging infection. This study investigate the influences of virulence properties of UPEC isolates with reference to multilocus sequence typing (MLST). The aim of this study was targeted that investigation of the bacterial pathogenicity associated with UTI in children. A total of 58 UPEC isolates were collected from urine samples from patients with clinical diagnosis of uncomplicated UTI. The MLST of UPEC strains were assessed by methods based on polymerase chain reaction. Motility was evaluated using soft-agar plates. Biofilm formation was analyzed in microtiter dish biofilm formation assay. Cell death assay was analyzed by Annexin V/Phosphatidylserine staining and DNA fragmentation assay. According the result, the predominant sequence type (ST) was ST95 (24.1%) and ST73 (17.2%). There were some difference in virulence gene and antibiotics resistance between ST95 and ST73. The number of 11 (18.9%) isolates were strongly adherent. Based on the detected biofilm formation, these strongly adherent are almost ST73. The ST95 was higher than ST73 in population, but ST95 was lower than ST73 in motility and cell death induction. This study indicated that the UPEC molecular strains are related to some virulence traits. Furthermore, the virulence factors carried by ST73 strains contribute to their abilities to colonize the host and cause disease.
Go to : 
REFERENCES
1). Asadi S, Kargar M, Solhjoo K, Najafi A, Ghorbani-Dalini S. The Association of Virulence Determinants of Uropathogenic Escherichia coli With Antibiotic Resistance. Jundishapur J Microbiol. 2014; 7:e9936.

2). Tabasi M, Asadi Karam MR, Habibi M, Yekaninejad MS, Bouzari S. Phenotypic Assays to Determine Virulence Factors of Uropathogenic Escherichia coli (UPEC) Isolates and their Correlation with Antibiotic Resistance Pattern. Osong Public Health Res Perspect. 2015; 6:261–8.
3). Blango MG, Mulvey MA. Persistence of uropathogenic Escherichia coli in the face of multiple antibiotics. Antimicrob Agents Chemother. 2010; 54:1855–63.
4). Lee S, Yu JK, Park K, Oh EJ, Kim SY, Park YJ. Phylogenetic groups and virulence factors in pathogenic and commensal strains of Escherichia coli and their association with blaCTX-M. Ann Clin Lab Sci. 2010; 40:361–7.
5). Bashir S, Sarwar Y, Ali A, Mohsin M, Saeed MA, Tariq A, et al. Multiple drug resistance patterns in various phylogenetic groups of uropathogenic E. coli isolated from Faisalabad region of Pakistan. Braz J Microbiol. 2011; 42:1278–83.
6). Obata-Yasuoka M, Ba-Thein W, Tsukamoto T, Yoshikawa H, Hayashi H. Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogeny with other extraintestinal E. coli. Microbiology. 2002; 148:2745–52.
7). Tartof SY, Solberg OD, Manges AR, Riley LW. Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol. 2005; 43:5860–4.
8). Banerjee R, Johnston B, Lohse C, Chattopadhyay S, Tchesnokova V, Sokurenko EV, et al. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob Agents Chemother. 2013; 57:5912–7.
9). Yun KW, Kim DS, Kim W, Lim IS. Molecular typing of uropathogenic Escherichia coli isolated from Korean Children with urinary tract infection. Korean J Pediatr. 2015; 58:20–7.
10). Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev. 1991; 4:80–128.
11). Le Bouguenec C, Garcia MI, Ouin V, Desperrier JM, Gounon P, Labigne A. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect Immun. 1993; 61:5106–14.
12). Lacher DW, Steinsland H, Whittam TS. Allelic subtyping of the intimin locus (eae) of pathogenic Escherichia coli by fluorescent RFLP. FEMS Microbiol Lett. 2006; 261:80–7.
13). Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998; 282:1494–7.
14). Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008; 85:11–9.
15). Kammler M, Schön C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993; 175:6212–9.
16). Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004; 2:123–40.
17). Uhlich GA, Cooke PH, Solomon EB. Analyses of the red-dry-rough phenotype of an Escherichia coli O157: H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol. 2006; 72:2564–72.
18). Nolan LM, Cavaliere R, Turnbull L, Whitchurch CB. Extra-cellular ATP inhibits twitching motility-mediated biofilm expansion by Pseudomonas aeruginosa. BMC Microbiol. 2015; 15:55.

19). Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016; 62:84–90.
20). Novais Â, Vuotto C, Pires J, Montenegro C, Donelli G, Coque TM, et al. Diversity and biofilm-production ability among isolates of Escherichia coli phylogroup D belonging to ST69, ST393 and ST405 clonal groups. BMC Microbiol. 2013; 13:144.
21). Mills M, Meysick KC, O'Brien AD. Cytotoxic necrotizing factor type 1 of uropathogenic Escherichia coli kills cultured human uroepithelial 5637 cells by an apoptotic mechanism. Infect Immun. 2000; 68:5869–80.
22). Dhakal BK, Mulvey MA. The UPEC pore-forming toxin alpha-hemolysin triggers proteolysis of host proteins to disrupt cell adhesion, inflammatory, and survival pathways. Cell Host Microbe. 2012; 11:58–69.
23). Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998; 391:43–50.

24). Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000; 181:261–72.
25). Gordon DM, Clermont O, Tolley H, Denamur E. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ Microbiol. 2008; 10:2484–96.
26). Emody L, Kerényi M, Nagy G. Virulence factors of uropathogenic Escherichia coli. Int J Antimicrob Agents. 2003; 22(Suppl 2):29–33.
27). Soto SM, Smithson A, Martinez JA, Horcajada JP, Mensa J, Vila J. Biofilm formation in uropathogenic Escherichia coli strains: relationship with prostatitis, urovirulence factors and antimicrobial resistance. J Urol. 2007; 177:365–8.
Go to : 
 | Figure 1.Biofilm formation of UPEC isolates. UPEC isolates incubate for 24 h at 37°C in LB broth without NaCl. Biofilm was stained by 0.1% crystal violet. Error bars represent the standard deviations of the results from three independent experiments. |
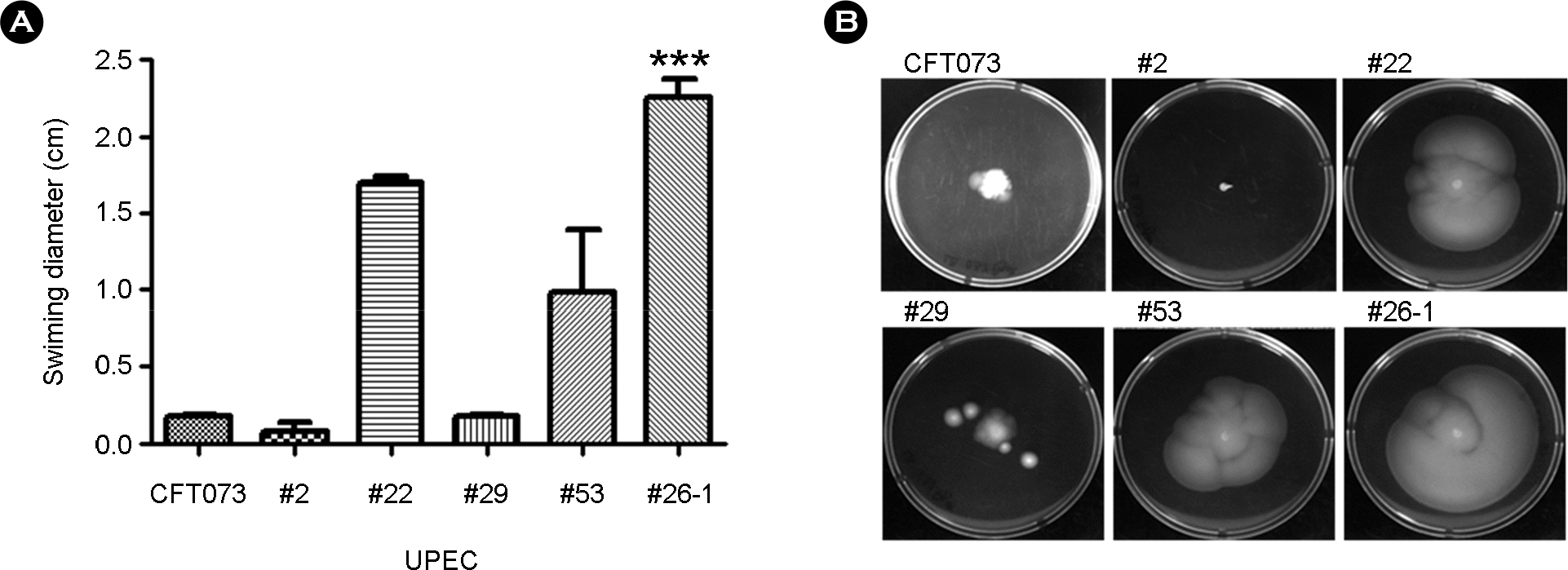 | Figure 2.Motility of UPEC. (A and B) Motility on 0.3% LB Agar of CFT073, isolated UPEC No.2, 22, 26-1, 53 and 29 after 16 h of incubation at 37°C. The errors bars in panel A represent the standard deviations from four independent experiments. Significant difference in motility was to determined using one-way ANOVA; ∗, p < 0.005. |
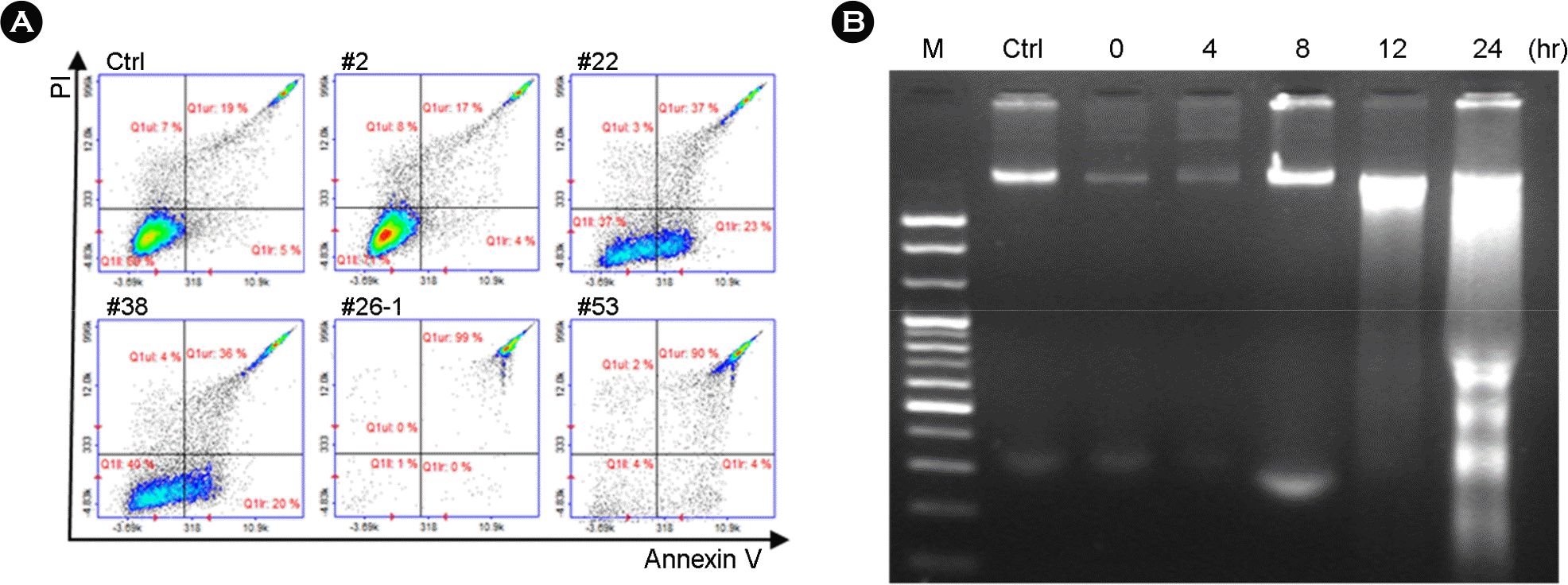 | Figure 3.UPEC-induced cell death. (A) Apoptosis of T24 infected with UPEC No. 2, 22, 38, 26-1 or 53 (MOI 10) for 3 h was measured by flow cytometry using Annexin V/PI staining. (B) DNA fragmentation analysis of T24 cells infected with UPEC isolate No. 26-1. T24 cells were infected with UPEC isolate No. 26-1 at an MOI of 10 for different time point at 37°C. M, 100 bp DNA marker; Ctrl, no infection. |
Table 1.
Primers used for MLST
Table 2.
Prevalence of sequence type and clonal complex in UPEC isolated from UTI
Table 3.
Prevalence of virulence gene in clonal complex
Table 4.
Prevalence of antibiotics resistance in clonal complex




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download