Abstract
Autophagy is a self-degradative process that removes misfolded or aggregated proteins, clears damaged organelles, as well as eliminates intracellular pathogens playing a role in innate immunity. Mycobacterium abscessus (M. abscessus) has been reported as a causative organism in nearly 80% of the rapid growing mycobacteria (RGM) pulmonary disease. The strain exhibits two different colony types: the smooth (S) one which is considered wild-type and the rough (R) one which is the mutated strain. In accordance to the colony morphology, the S and R types display varying autophagic responses in the host cells with the R type inducing elevated autophagy compared to the S type. The major difference in the autophagy could be based on the bioactive molecules exposed on the surface of the S and R types. Though autophagy has a vital role to play in the clearance of intracellular pathogens, very little is known on the autophagy induced by M. abscessus. It has been known that the intracellular pathogens employ different strategies to evade the autophagic pathway and to survive within the host cells. This review summarizes the most up-to-date findings on autophagy induced by M. abscessus morphotypes and how M. abscessus evades the autophagic machinery to divide and thrive inside the host cells. In addition, the prospects of autophagic machinery in devising new anti-infective strategies against mycobacterial infection is also been discussed.
Go to : 
REFERENCES
1). Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004; 14:70–7.
2). Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005; 118:7–18.

3). Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004; 6:463–77.
4). Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007; 27:19–40.

5). Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004; 306:990–5.

6). Ryter SW, Cloonan SM, Choi AM. Autophagy: a critical regulator of cellular metabolism and homeostasis. Mol Cells. 2013; 36:7–16.

7). Nowikovsky K, Devenish RJ, Froschauer E, Schweyen RJ. Determination of yeast mitochondrial KHE activity, osmotic swelling and mitophagy. Methods Enzymol. 2009; 457:305–17.
8). Walter KM, Schönenberger MJ, Trötzmüller M, Horn M, Elsässer HP, Moser AB, et al. Hif-2α promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab. 2014; 20:882–97.

10). Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. J Clin Invest. 2015; 125:65–74.

12). Gozuacik D, Kimchi A. Autophagy as a cell death and tumor suppressor mechanism. Oncogene. 2004; 23:2891–906.

13). Netea-Maier RT, Plantinga TS, van de Veerdonk FL, Smit JW, Netea MG. Modulation of inflammation by autophagy: Consequences for human disease. Autophagy. 2016; 12:245–60.

15). Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al. Autophagosome formation from membrane compartments enriched in phosphatidylnositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008; 182:685–701.
16). Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009; 186:773–82.

17). Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009; 10:458–67.

18). Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007; 8:931–7.

19). Kundu M, Thompson CB. Macroautophagy versus mitochondrial autophagy: a question of fate? Cell Death Differ. 2005; 12(Suppl 2):1484–9.

21). Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009; 34:259–69.

23). Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol. 2005; 167:1713–28.

24). Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann-Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000; 406:902–6.

25). Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007; 6:304–12.

26). Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, et al. Aggregate-prone proteins are cleared from the cytosol by autophagy: therapeutic implications. Curr Top Dev Biol. 2006; 76:89–101.

27). Perlmutter DH. The role of autophagy in alpha-1-antitrypsin deficiency: a specific cellular response in genetic diseases associated with aggregation-prone proteins. Autophagy. 2006; 2:258–63.

28). Endo Y, Furuta A, Nishino I. Danon disease: a phenotypic expression of LAMP-2 deficiency. Acta Neuropathol. 2015; 129:391–8.

29). Terman A, Brunk UT. Autophagy in cardiac myocyte homeostasis, aging, and pathology. Cardiovasc Res. 2005; 68:355–65.

30). Levine B. Cell biology: autophagy and cancer. Nature. 2007; 446:745–7.
31). Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007; 7:961–7.

32). Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991; 253:49–53.

33). Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999; 402:672–6.

34). Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006; 8:1124–32.

35). Mariño G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, López-Otín C. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007; 282:18573–83.

36). Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006; 8:688–99.

37). Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007; 9:1142–51.

38). Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013; 13:722–37.

39). Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat Rev Microbiol. 2014; 12:101–14.

40). Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009; 5:527–49.

41). Walker DH, Popov VL, Crocquet-Valdes PA, Welsh CJ, Feng HM. Cytokine-induced, nitric oxide-dependent, intracellular antirickettsial activity of mouse endothelial cells. Lab Invest. 1997; 76:129–38.
42). Vergne I, Chua J, Singh SB, Deretic V. Cell biology of mycobacterium tuberculosis phagosome. Annu Rev Cell Dev Biol. 2004; 20:367–94.

43). Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004; 119:753–66.
44). Castillo EF, Dekonenko A, Arko-Mensah J, Mandell MA, Dupont N, Jiang S, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci U S A. 2012; 109:E3168–76.

45). Brumell JH, Grinstein S. Salmonella redirects phagosomal maturation. Curr Opin Microbiol. 2004; 7:78–84.
46). Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006; 281:11374–83.
47). Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004; 306:1037–40.
48). Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005; 307:727–31.
49). Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 2006; 4:423–34.
50). Shin DM, Jeon BY, Lee HM, Jin HS, Yuk JM, Song CH, et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010; 6:e1001230.
51). Horwitz MA. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J Exp Med. 1983; 158:2108–26.
52). Howe D, Mallavia LP. Coxiella burnetii exhibits morphological change and delays phagolysosomal fusion after internalization by J774A.1 cells. Infect Immun. 2000; 68:3815–21.
53). Proctor RA, Balwit JM, Vesga O. Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect Agents Dis. 1994; 3:302–12.
54). Ogawa M, Sasakawa C. Shigella and autophagy. Autophagy. 2006; 2:171–4.
55). Goren MB. Phagocyte lysosomes: interactions with infectious agents, phagosomes, and experimental perturbations in function. Annu Rev Microbiol. 1977; 31:507–33.

56). Gorden AH, Hart PA, Young MR. Ammonia inhibits phagosomes-lysosome fusion in macrophages. Nature. 1980; 286:79–80.
57). Sturgill-Koszycki S, Schlesinger PH, Chakraborty P, Haddix PL, Collins HL, Fok AK, et al. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994; 263:678–81.
58). Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001; 154:631–44.

59). Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004; 53:29–40.
60). Howe D, Melnicákova J, Barák I, Heinzen RA. Fusogenicity of the Coxiella burnetii parasitophorous vacuole. Ann N Y Acad Sci. 2003; 990:556–62.
61). Baca OG, Li YP, Kumar H. Survival of the Q fever agent Coxiella burnetii in the phagolysosome. Trends Microbiol. 1994; 2:476–80.
62). Baca OG, Paretsky D. Q fever and Coxiella burnetii: a model for host-parasite interactions. Microbiol Rev. 1983; 47:127–49.
63). Delgado M, Singh S, De Haro S, Master S, Ponpuak M, Dinkins C, et al. Autophagy and pattern recognition receptors in innate immunity. Immunol Rev. 2009; 227:189–202.

64). Deretic V. Autophagy as an innate immunity paradigm: expanding the scope and repertoire of pattern recognition receptors. Curr Opin Immunol. 2012; 24:21–31.

66). Perot BP, Ingersoll MA, Albert ML. The impact of macroautophagy on CD8 (+) T-cell-mediated antiviral immunity. Immunol Rev. 2013; 255:40–56.
67). Brown-Elliott BA, Wallace RJ Jr. Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev. 2002; 15:716–46.

68). Griffith DE, Girard WM, Wallace RJ Jr. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis. 1993; 147:1271–8.

69). Sanguinetti M, Ardito F, Fiscarelli E, La Sorda M, D'Argenio P, Ricciotti G, et al. Fatal pulmonary infection due to multidrug-resistant Mycobacterium abscessus in a patient with cystic fibrosis. J Clin Microbiol. 2001; 39:816–9.
70). Jeon K, Kwon OJ, Lee NY, Kim BJ, Kook YH, Lee SH, et al. Antibiotic treatment of Mycobacterium abscessus lung disease: a retrospective analysis of 65 patients. Am J Respir Crit Care Med. 2009; 180:896–902.
71). Chatterjee D, Khoo KH. The surface glycopeptidolipids of mycobacteria: structures and biological properties. Cell Mol Life Sci. 2001; 58:2018–42.

72). Howard ST, Rhoades E, Recht J, Pang X, Alsup A, Kolter R, et al. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology. 2006; 152:1581–90.
73). Fregnan GB, Smith DW. Description of various colony forms of mycobacteria. J Bacteriol. 1962; 83:819–27.

74). Eckstein TM, Inamine JM, Lambert ML, Belisle JT. A genetic mechanism for deletion of the ser2 gene cluster and formation of rough morphological variants of Mycobacterium avium. J Bacteriol. 2000; 182:6177–82.
75). Barrow WW, Brennan PJ. Isolation in high frequency of rough variants of Mycobacterium intracellulare lacking C-mycoside glycopeptidolipid antigens. J Bacteriol. 1982; 150:381–4.
76). Rottman M, Catherinot E, Hochedez P, Emile JF, Casanova JL, Gaillard JL, et al. Importance of T cells, gamma interferon, and tumor necrosis factor in immune control of the rapid grower Mycobacterium abscessus in C57BL/6 mice. Infect Immun. 2007; 75:5898–907.
77). Sohn H, Kim HJ, Kim JM, Jung Kwon O, Koh WJ, Shin SJ. High virulent clinical isolates of Mycobacterium abscessus from patients with the upper lobe fibrocavitary form of pulmonary disease. Microb Pathog. 2009; 47:321–8.
78). Rhoades ER, Archambault AS, Greendyke R, Hsu FF, Streeter C, Byrd TF. Mycobacterium abscessus Glycopeptidolipids mask underlying cell wall phosphatidyl-myo-inositol mannosides blocking induction of human macrophage TNF-alpha by preventing interaction with TLR2. J Immunol. 2009; 183:1997–2007.
79). Jönsson BE, Gilljam M, Lindblad A, Ridell M, Wold AE, Welinder-Olsson C. Molecular epidemiology of Mycobacterium abscessus, with focus on cystic fibrosis. J Clin Microbiol. 2007; 45:1497–504.
80). Roux AL, Viljoen A, Bah A, Simeone R, Bernut A, Laencina L, et al. The distinct fate of smooth and rough Mycobacterium abscessus variants inside macrophages. Open Biol. 2016; 6.
81). Bernut A, Herrmann JL, Kissa K, Dubremetz JF, Gaillard JL, Lutfalla G, et al. Mycobacterium abscessus cording prevents phagocytosis and promotes abscess formation. Proc Natl Acad Sci USA. 2014; 111:E943–52.
82). Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity. 2010; 32:329–41.

83). Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009; 137:1001–4.

84). Zullo AJ, Lee S. Mycobacterial Induction of Autophagy Varies by Species and Occurs Independently of Mammalian Target of Rapamycin Inhibition. J Biol Chem. 2012; 287:12668–78.

85). Pizarro-Cerdá J, Moreno E, Sanguedolce V, Mege JL, Gorvel JP. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun. 1998; 66:2387–92.
86). Rao V, Fujiwara N, Porcelli SA, Glickman MS. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J Exp Med. 2005; 201:535–43.
87). Saitoh T, Akira S. Regulation of innate immune responses by autophagy-related proteins. J Cell Biol. 2010; 189:925–35.

88). Forrellad MA, Klepp LI, Gioffré A, Sabio y García J, Morbidoni HR, de la Paz Santangelo M, et al. Virulence factors of the Mycobacterium tuberculosis complex. Virulence. 2013; 4:3–66.
Go to : 
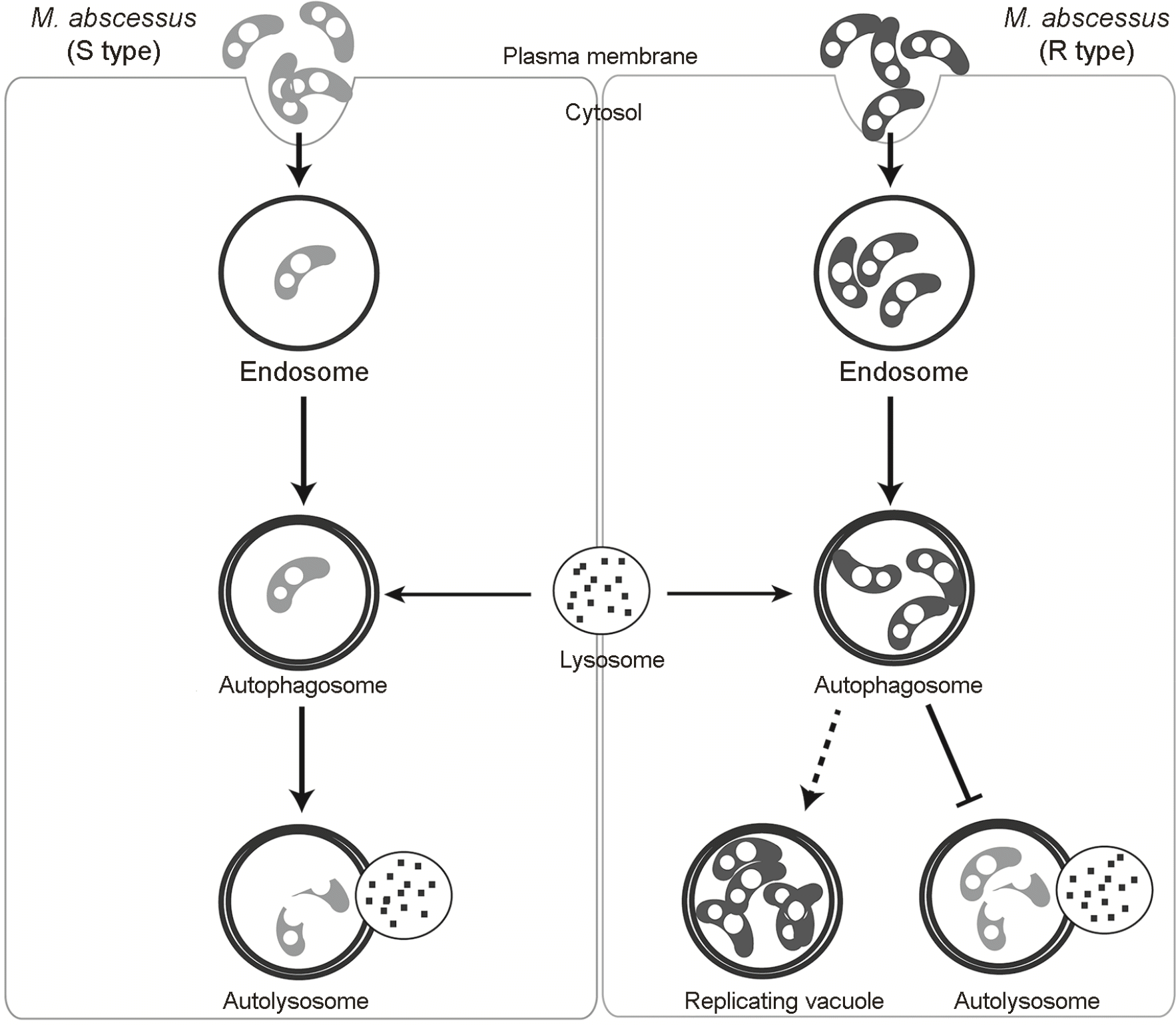 | Figure 1.Proposed model of the autophagic pathway in response to S and R type M. abscessus infiltration. After invasion, the bacteria are contained in endosomes. The R type endosomes are characterized by the presence of multiple bacterial cells in contrast to the S type. The endosomes fuse with early autophagosomes which later matures into late autophagosomes. The late autophagosomes eventually fuse with the lysosomes to become autolysosomes in which the bacteria are killed. The R type M. abscessus inhibits lysosomal fusion and replicate within replicating vacuoles that resemble late autophagosomes. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download