Abstract
Orientia tsutsugamushi (O. tsutsugamushi), which is endemic to an Asia-Pacific region, has increased its incidence and caused annually around 10 thousand patients infected with scrub typhus in Korea in the past several years. In the present study, we isolated 44 O. tsutsugamushi from the patients with febrile illness accompanied with or without an eschar in Gyeongsangnam-do, Korea. These isolates were characterized by genetic analysis of the major outer membrane protein, the 56-kDa type-specific antigen (tsa56), which is unique to O. tsutsugamushi. Two types of sequences of tsa56, designated by JJ1 and JJ2, were determined from 37 and 7 isolates of the 44 isolates, respectively. JJ1 and JJ2 showed 74.7~90.8% identity in nucleotide sequence and 66.1~90.5% identity in amino acid sequence with 33 reference strains except for Boryong and Kuroki. JJ1 and JJ2 had 100 and 99.9% nucleotide identity to Boryong strain, and 99.9 and 99.8 % to Kuroki, which has been known to be similar to Boryong, respectively. In addition, they showed 77.9~ 81.4% nucleotide identity with the cluster of Gilliam-related genotypes, whereas they showed higher nucleotide identity (89.6~90.8%) with the cluster of Karp-related genotypes. To our knowledge, this is the first report to isolate O. tsutsugamushi and characterize their genotype as the Boryong in Jinju and West Gyeongsangnam-do, Korea, even though it has been reported that the Boryong was the predominant genotype in isolates from chiggers, domestic rodents, and patients in the southern part of Korea. Furthermore, our isolates could be useful source to study on the pathophysiology and epidemiology of scrub typhus in Korea.
Go to : 
REFERENCES
1). Tamura A, Ohashi N, Urakami H, Miyamura S. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int J Syst Bacteriol. 1995; 45:589–91.
2). Kim DM, Yun NR, Neupane GP, Shin SH, Ryu SY, Yoon HJ, et al. Differences in clinical features according to Boryoung and Karp genotypes of Orientia tsutsugamushi. PLoS One. 2011; 6:e22731.
3). Yang HH, Huang IT, Lin CH, Chen TY, Chen LK. New genotypes of Orientia tsutsugamushi isolated from humans in Eastern Taiwan. PLoS One. 2012; 7:e46997.
4). Nakayama K, Yamashita A, Kurokawa K, Morimoto T, Ogawa M, Fukuhara M, et al. The Whole-genome sequencing of the obligate intracellular bacterium Orientia tsutsugamushi revealed massive gene amplification during reductive genome evolution. DNA Res. 2008; 15:185–99.
5). Shirai A, Tanskul PL, Andre RG, Dohany AL, Huxsoll DL. Rickettsia tsutsugamushi strains found in chiggers collected in Thailand. Southeast Asian J Trop Med Public Health. 1981; 12:1–6.
6). Ree HI, Kim TE, Lee IY, Jeon SH, Hwang UW, Chang WH. Determination and geographical distribution of Orientia tsutsugamushi serotypes in Korea by nested polymerase chain reaction. Am J Trop Med Hyg. 2001; 65:528–34.
7). Zhang M, Zhao ZT, Yang HL, Zhang AH, Xu XQ, Meng XP, et al. Molecular epidemiology of Orientia tsutsugamushi in chiggers and ticks from domestic rodents in Shandong, northern China. Parasit Vectors. 2013; 6:312.

8). Kelly DJ, Fuerst PA, Ching WM, Richards AL. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis. 2009; 48(Suppl 3):S203–30.
9). Chang WH, Kang JS, Lee WK, Choi MS, Lee JH. Serological classification by monoclonal antibodies of Rickettsia tsutsugamushi isolated in Korea. J Clin Microbiol. 1990; 28:685–8.
10). Yamashita T, Kasuya S, Noda N, Nagano I, Kang JS. Transmission of Rickettsia tsutsugamushi strains among humans, wild rodents, and trombiculid mites in an area of Japan in which tsutsugamushi disease is newly endemic. J Clin Microbiol. 1994; 32:2780–5.
11). Furuya Y, Yoshida Y, Katayama T, Yamamoto S, Kawamura A Jr. Serotype-specific amplification of Rickettsia tsutsugamushi DNA by nested polymerase chain reaction. J Clin Microbiol. 1993; 31:1637–40.
12). Chong YS, Kim JJ, Kwak CG, Lee SY, Yi KS, Suto T. Isolation of Rickettsia tsutsugamushi from patients in Chinhae area in 1987. J Korean Soc Microbiol. 1989; 24:81–7.
13). Rodkvamtook W, Ruang-Areerate T, Gaywee J, Richards AL, Jeamwattanalert P, Bodhidatta D, et al. Isolation and characterization of Orientia tsutsugamushi from rodents captured following a scrub typhus outbreak at a military training base, Bothong district, Chonburi province, central Thailand. Am J Trop Med Hyg. 2011; 84:599–607.
14). Blacksell SD, Luksameetanasan R, Kalambaheti T, Aukkanit N, Paris DH, McGready R, et al. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol Med Microbiol. 2008; 52:335–42.
15). Bengston IA. A serological study of 37 cases of tsutsugamushi disease (scrub typhus) occurring in Burma and the Philippine Islands. Public Health Rep. 1946; 61:887–94.
16). Tamura A, Makisaka Y, Kadosaka T, Enatsu T, Okubo K, Koyama S, et al. Isolation of Orientia tsutsugamushi from Leptotrombidium fuji and its characterization. Microbiol Immunol. 2000; 44:201–4.
17). Tamura A, Takahashi K, Tsuruhara T, Urakami H, Miyamura S, Sekikawa H, et al. Isolation of Rickettsia tsutsugamushi antigenically different from Kato, Karp, and Gilliam strains from patients. Microbiol Immunol. 1984; 28:873–82.
18). Derrick EH, Brown HE. Isolation of the Karp strain of rickettsia tsutsugamushi. Lancet. 1949; 2:150.

19). Shishido A, Ohtawara M, Hikita M, Kitaoka M. The nature of immunity against scrub typhus in mice. II. The cross-protection test with mice for identification and differentiation of several strains of Rickettsia orientalis newly isolated in Japan. Jpn J Med Sci Biol. 1959; 12:391–404.
20). Yamamoto S, Kawabata N, Tamura A, Urakami H, Ohashi N, Murata M, et al. Immunological properties of Rickettsia tsutsugamushi, Kawasaki strain, isolated from a patient in Kyushu. Microbiol Immunol. 1986; 30:611–20.
21). Ohashi N, Tamura A, Sakurai H, Yamamoto S. Chaacterization of a new antigenic type, Kuroki, of Rickettsia tsutsugamushi isolated from a patient in Japan. J Clin Microbiol. 1990; 28:2111–3.
22). Enatsu T, Urakami H, Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol Lett. 1999; 180:163–9.
23). Shirai A, Robinson DM, Brown GW, Gan E, Huxsoll DL. Antigenic analysis by direct immunofluorescence of 114 isolates of Rickettsia tsutsugamushi recovered from febrile patients in rural Malaysia. Jpn J Med Sci Biol. 1979; 32:337–44.
24). Qiang Y, Tamura A, Urakami H, Makisaka Y, Koyama S, Fukuhara M, et al. Phylogenetic characterization of Orientia tsutsugamushi isolated in Taiwan according to the sequence homologies of 56-kDa type-specific antigen genes. Microbiol Immunol. 2003; 47:577–83.
Go to : 
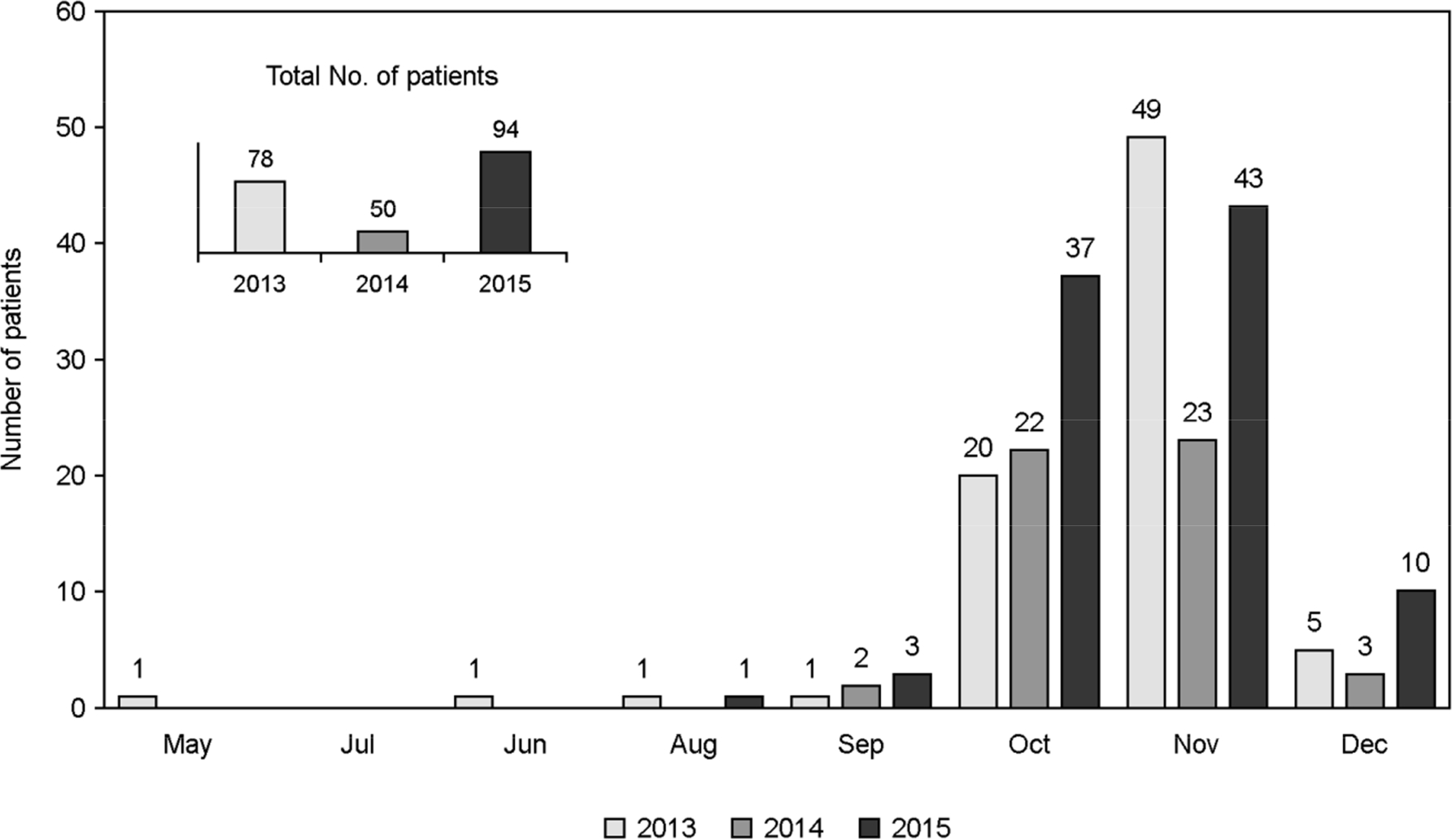 | Figure 1.The incidence of the scrub typhus patients of Gyeongsang National University Hospital in 2013 to 2015. |
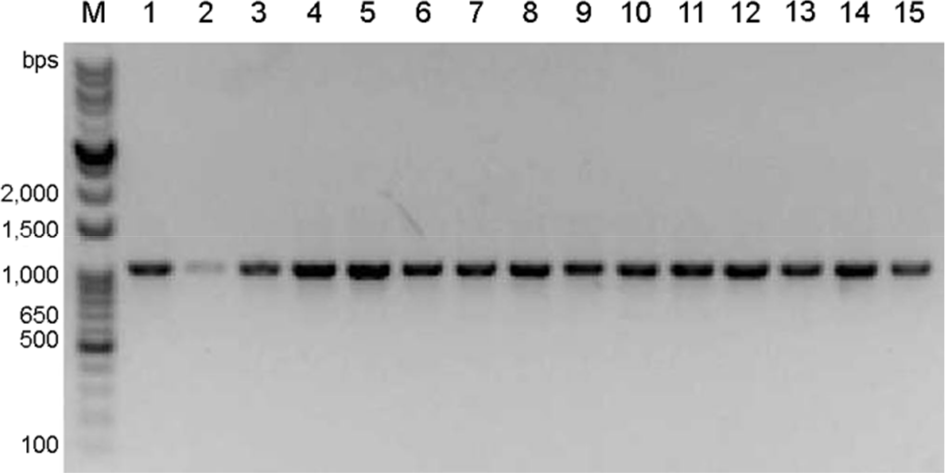 | Figure 2.The PCR products of the 56-kDa type-specific antigen gene of O. tsutsugamushi from 15 representative isolates in the present study. Forty-four O. tsutsugamushi isolates were confirmed by tsa56-specific PCR assay, and PCR products of 15 representative isolates among 44 isolates were presented. M, DNA sizer marker. |
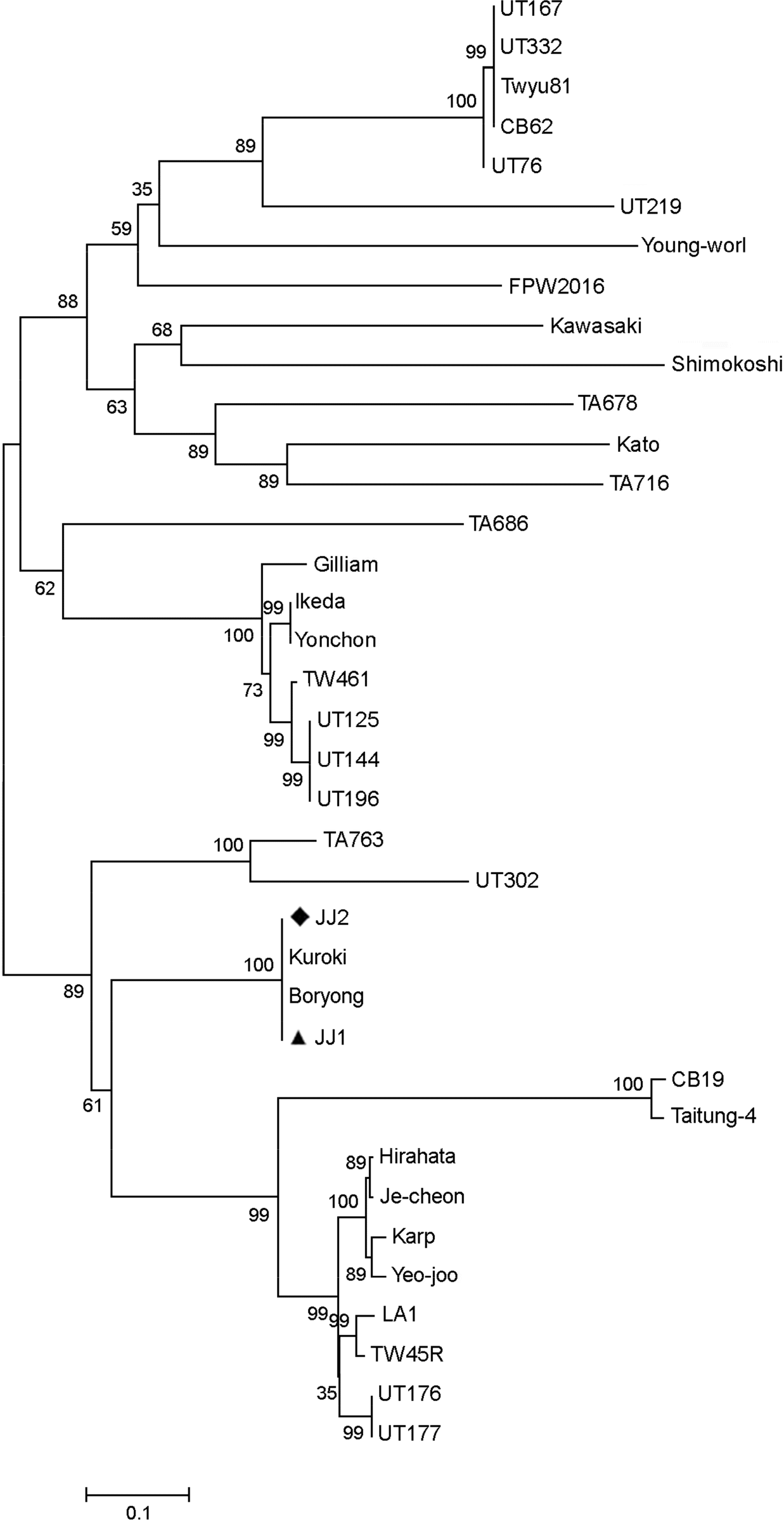 | Figure 3.Phylogenetic tree based on partial 56-kDa type specific antigen of O. tsutsugamushi. Two nucleotide sequences of O. tsutsugamushi isolates were analyzed phylogenetic relationship with reference sequences retrieved from the GenBank database as shown in Table 1. Solid triangle and diamond indicate two sequences of O. tsutsugamushi determined in this study. |
Table 1.
Sequence homologies of partial 56-kDa type specific antigen gene between Orientia tsutsugamushi strains
| Isolates | Nucleotide identity (%) | Amino acid identity (%) | Information | ||||||
|---|---|---|---|---|---|---|---|---|---|
| JJ1 | JJ2 | JJ1 | JJ2 | Source | Country | Year | Accession no. | Reference | |
| Boryong | 100 | 99.9 | 100 | 100 | Human | South Korea | 1998 | L04956 | (9) |
| CB19 | 77.9 | 78.1 | 68.1 | 68.1 | Rattus rattus | Thailand | 2009 | GU068058 | (13) |
| CB62 | 85.8 | 85.6 | 78.3 | 78.3 | Bandicota indica | Thailand | 2009 | GU068055 | (13) |
| FPW2016 | 78.1 | 77.9 | 67 | 67 | Human | Thailand | 2007 | EF213085 | (14) |
| Gilliam∗ | 86.5 | 86.4 | 78.3 | 78.3 | Human | Assam-Burma border | 1943 | DQ485289 | (15) |
| Hirahata | 90.8 | 90.7 | 90.5 | 90.5 | Leptotrombidium pallidum | Taiwan | 1999 | AF201835 | (16) |
| Ikeda | 81.2 | 81.1 | 72.3 | 72.3 | Human | Japan | 1979 | AF173033 | (17) |
| Je-cheon | 90.7 | 90.6 | 84.2 | 84.2 | Human | South Korea | 2001 | AF430143 | – |
| Karp∗ | 90.6 | 90.5 | 84.6 | 84.6 | Human | New Guinea | 1943 | AY956315 | (18) |
| Kato∗ | 76.9 | 76.8 | 66.1 | 66.1 | Human | Japan | 1955 | AY836148 | (19) |
| Kawasaki | 78.0 | 77.9 | 66.5 | 66.5 | Human | Japan | 1981 | M63383 | (20) |
| Kuroki | 99.9 | 99.8 | 100 | 100 | Human | Japan | 1981 | M63380 | (21) |
| LA-1 | 90.0 | 89.9 | 82.8 | 82.8 | Leptotrombidium arenicola | Malaysia | 1993 | AF173049 | (22) |
| LF-1 | 76.2 | 76.1 | 66.8 | 66.8 | Leptotrombidium fletcheri | Malaysia | 1993 | AF173050 | (22) |
| Shimokoshi | 74.8 | 74.7 | 61.4 | 61.4 | Human | Japan | 1980 | M63381 | (17) |
| TA678 | 78.1 | 78.2 | 67 | 67 | Rattus rattus | Thailand | 1963 | U19904 | (23) |
| TA686 | 82.0 | 82.1 | 73.1 | 73.1 | Tupaia glis | Thailand | 1963 | U80635 | (23) |
| TA716 | 78.3 | 78.4 | 67.8 | 67.8 | Menetes berdmorei | Thailand | 1963 | U19905 | (23) |
| TA763 | 84.1 | 84.2 | 74.6 | 74.6 | Rattus rajah | Taiwan | 1963 | U80636 | (23) |
| Taitung-4 | 78.9 | 79.0 | 70.2 | 70.2 | – | Taiwan | 2004 | AY787232 | – |
| TW45R | 90.7 | 90.6 | 84.2 | 84.2 | Rattus losea | Taiwan | 2003 | AY222632 | (24) |
| TW461 | 81.1 | 81.0 | 71 | 71 | Rattus rattus | Taiwan | 2003 | AY222631 | (24) |
| TWyu81 | 86.1 | 86.0 | 77.9 | 77.9 | Leptotrombidium pallidum | Thailand | 2003 | AY222640 | (24) |
| UT76 | 85.9 | 85.8 | 71 | 71 | Human | Thailand | 2007 | EF213078 | (14) |
| UT125 | 81.4 | 81.2 | 71 | 71 | Human | Thailand | 2007 | EF213096 | (14) |
| UT144 | 81.4 | 81.2 | 78.3 | 78.3 | Human | Thailand | 2007 | EF213091 | (14) |
| UT167 | 86.1 | 86.0 | 80.6 | 80.6 | Human | Thailand | 2007 | EF213080 | (14) |
| UT176 | 89.8 | 89.6 | 80.6 | 80.6 | Human | Thailand | 2007 | EF213081 | (14) |
| UT177 | 89.8 | 89.6 | 71 | 71 | Human | Thailand | 2007 | EF213084 | (14) |
| UT196 | 81.4 | 81.2 | 86.3 | 86.3 | Human | Thailand | 2007 | EF213079 | (14) |
| UT219 | 90.6 | 90.5 | 69.5 | 69.5 | Human | Thailand | 2007 | EF213100 | (14) |
| UT302 | 79.4 | 79.6 | 78.3 | 78.3 | Human | Thailand | 2007 | EF213095 | (14) |
| UT332 | 86.1 | 86.0 | 77.9 | 77.9 | Human | Thailand | 2007 | EF213083 | (14) |
| Yeojoo | 90.4 | 90.2 | 84.2 | 84.2 | Human | South Korea | 2001 | AF430144 | – |
| Yonchon | 81.2 | 81.1 | 72.1 | 72.1 | Human | South Korea | 1989 | U19903 | (25) |
| Young-worl | 87.8 | 87.7 | 79.4 | 79.4 | Human | South Korea | 2001 | AF430141 | – |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download