Abstract
Community-associated methicillin resistant Staphylococcus aureus (CA-MRSA) has become widespread in the community and healthcare settings, and a number of clonal lineages emerged on every country. Sequence type (ST) 80 clone of CA-MRSA was dominant in Europe and has increasingly been isolated from the Middle East but so far never found in Korea. In this study, 48 MRSA isolates recovered from ear infections were characterized by multilocus sequence typing (MLST), staphylococcal cassette chromosome mec (SCC mec) typing, staphylocoagulase (SC) genotyping, staphylococcal protein A gene (spa) typing, accessory gene regulator (agr) typing, and virulence gene profiling. Most MRSA strains belonged to three major clones: ST5-SCC mec II-SC type II (n=19, 39.6%), ST239-SCC mec III-SC type IV (n=15, 31.2%), and ST72-SCC mec IV-SC type Vb (n=11, 22.9%). Among the isolates, one strain was Panton-Valentine leukocidin (PVL)-positive ST80-SCC mec IV-SC type XIa – spa type t044-agr group III, and exfoliative toxin D-positive. This strain was susceptible to most antibiotics, but resistant to tetracycline and fusidic acid. This is the first report on the emergence of European ST80 CA-MRSA clone in Korea.
Go to : 
REFERENCES
1). Chen CJ, Huang YC. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect. 2014; 20:605–23.
2). Fluit AC, Carpaij N, Majoor EA, Weinstein RA, Aroutcheva A, Rice TW, et al. Comparison of an ST80 MRSA strain from the USA with European ST80 strains. J Antimicrob Chemother. 2015; 70:664–9.

3). Ben Nejma M, Mastouri M, Bel Hadj Jrad B, Nour M. Characterization of ST80 Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus clone in Tunisia. Diagn Microbiol Infect Dis. 2013; 77:20–4.
4). Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States Hospitals. Antimicrob Agents Chemother. 2012; 56:1324–30.
5). Chuang YY, Huang YC. Molecular epidemiology of community-associated meticillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. 2013; 13:698–708.
6). Bae IG, Kim JS, Kim S, Heo ST, Chang C, Lee EY. Genetic correlation of community-associated methicillin-resistant Staphylococcus aureus strains from carriers and from patients with clinical infection in one region of Korea. J Korean Med Sci. 2010; 25:197–202.
7). Kim ES, Lee HJ, Chung GT, Lee YS, Shin DH, Jung SI, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus isolates in Korea. J Clin Microbiol. 2011; 49:1979–82.
8). Park SH, Kim KJ, Kim BK, Hwang SM. Molecular characterization of community-associated methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from children with skin infections in Busan, Korea. J Bacteriol Virol. 2015; 45:104–11.
9). Kim SY, Park SH, Hwang SM. Prevalence and molecular characteristics of methicillin-resistant Staphylococcus aureus isolates in a neonatal intensive care unit. J Bacteriol Virol. 2016; 46:99–103.
10). Ichiyama S, Ohta M, Shimokata K, Kato N, Takeuchi J. Genomic DNA fingerprinting by pulsed-field gel electrophoresis as an epidemiological marker for study of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1991; 29:2690–5.
11). Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000; 38:1008–15.
12). Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002; 46:2155–61.
13). Watanabe S, Ito T, Sasaki T, Li S, Uchiyama I, Kishii K, et al. Genetic diversity of staphylocoagulase genes (coa): insight into the evolution of variable chromosomal virulence factors in Staphylococcus aureus. PLoS One. 2009; 4:e5714.
14). Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, Vogel U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. 2003; 41:5442–8.
15). Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991; 29:2240–4.

16). Hirose M, Kobayashi N, Ghosh S, Paul SK, Shen T, Urushibara N, et al. Identification of staphylocoagulase genotypes I-X and discrimination of type IV and V subtypes by multiplex PCR assay for clinical isolates of Staphylococcus aureus. Jpn J Infect Dis. 2010; 63:257–63.
17). Shopsin B, Mathema B, Alcabes P, Said-Salim B, Lina G, Matsuka A, et al. Prevalence of agr specificity groups among Staphylococcus aureus strains colonizing children and their guardians. J Clin Microbiol. 2003; 41:456–9.
18). Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999; 29:1128–32.
19). Johnson WM, Tyler SD, Ewan EP, Ashton FE, Pollard DR, Rozee KR. Detection of genes for enterotoxins, exfoliative toxins, and toxic shock syndrome toxin 1 in Staphylococcus aureus by the polymerase chain reaction. J Clin Microbiol. 1991; 29:426–30.
20). Stegger M, Price LB, Larsen AR, Gillece JD, Waters AE, Skov R, et al. Genome sequence of Staphylococcus aureus strain 11819-97, an ST80-IV European community-acquired methicillin-resistant isolate. J Bacteriol. 2012; 194:1625–6.
21). Larsen AR, Stegger M, Bocher S, Sorum M, Monnet DL, Skov RL. Emergence and characterization of community-associated methicillin-resistant Staphyloccocus aureus infections in Denmark, 1999 to 2006. J Clin Microbiol. 2009; 47:73–8.
22). Watanabe S, Ito T, Takeuchi F, Endo M, Okuno E, Hiramatsu K. Structural comparison of ten serotypes of staphylocoagulases in Staphylococcus aureus. J Bacteriol. 2005; 187:3698–707.
23). Kanemitsu K, Yamamoto H, Takemura H, Kaku M, Shimada J. Relatedness between the coagulase gene 3′-end region and coagulase serotypes among Staphylococcus aureus strains. Microbiol Immunol. 2001; 45:23–7.
24). Hwang SM, Seki K, Sakurada J, Ogasawara M, Murai M, Ohmayu S, et al. Improved methods for detection and serotyping of coagulase from Staphylococcus aureus. Microbiol Immunol. 1989; 33:175–82.
25). Song Y, Du X, Li T, Zhu Y, Li M. Phenotypic and molecular characterization of Staphylococcus aureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010. J Med Microbiol. 2013; 62:274–82.
26). Park C, Lee DG, Kim SW, Choi SM, Park SH, Chun HS, et al. Predominance of community-associated methicillin-resistant Staphylococcus aureus strains carrying staphylococcal chromosome cassette mec type IVA in South Korea. J Clin Microbiol. 2007; 45:4021–6.
Go to : 
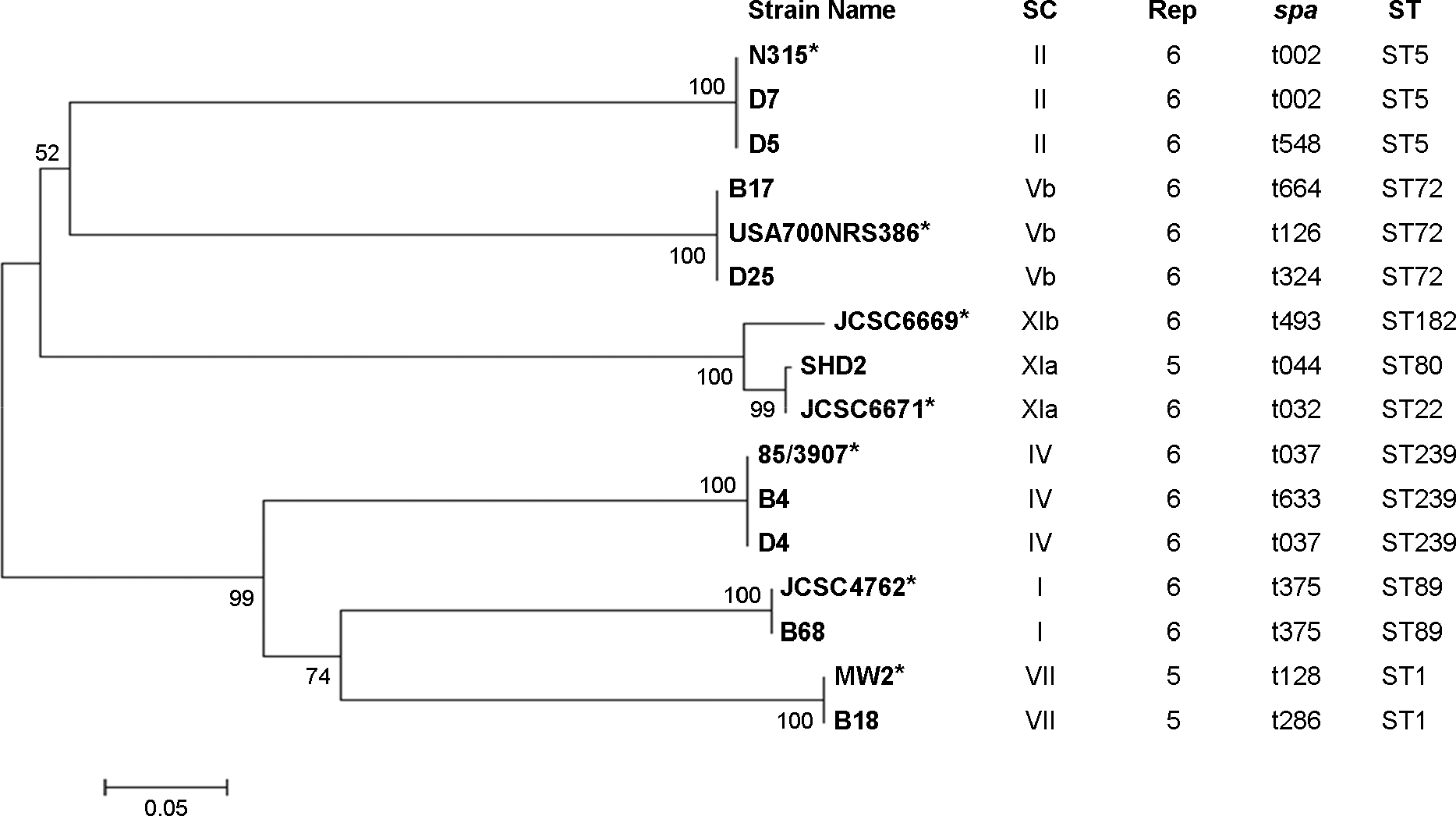 | Figure 1.Phylogenetic relationship among the nucleotide sequences of the D1 coa regions of the selected 9 isolates and 7 reference strains. The numbers at nodes indicate the bootstrap values based on 1000 replicates. ∗, reference strains; SC, staphylocoagulase type; Rep, a number of C-terminal repeat units of coa; spa, staphylococcal protein A gene; ST, sequence type. |
Table 1.
Molecular characteristics of 48 MRSA isolates from ear infections
| MLSTa (strain) | No. of isolates | SCC mec type | SCb type | spa type (n) | SSRc profile | agr type | Toxin genes se, tst, et (n) | PVL |
|---|---|---|---|---|---|---|---|---|
| se, tst, et (n) | ||||||||
| ST5 | 19 | II | II | t002(17) t548(2) | 26-23-17-34-17-20-17-12-17-16 26-23-17-34-17-20-17-12-16 | II | sec ∙ tst (14) tst (5) | - |
| ST239 | 15 | III | IV | t037(14) t633(1) | 15-12-16-02-25-17-24 08-24 | I | sea (6) | - |
| ST72 | 11 | IV | Vb | t324(6) t664(5) | 07-23-12-12-17-20-17-12-12-17 07-23-12-12-17-20-17-12-17 | I | - | - |
| ST80 (SHD2) | 1 | IV | XIa | t044 | 07-23-12-34-34-33-34 | III | etd | + |
| ST89 | 1 | II | I | t375 | 49-13-23-05-17-34-33-34 | III | etb | - |
| ST1 | 1 | IV | VII | t286 | 07-23-13-34-16-34-33-13 | III | - | - |
Table 2.
Antimicrobial resistance rate of 48 MRSA isolates from ear infections




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download