Abstract
Japanese encephalitis (JE) is a zoonosis that affects the nervous system of humans and other animals. The genotype of JE virus (JEV) has shifted recently from genotype 3 (G3) to genotype 1 (G1) in Asia, including Korea. Thus, a rapid differential assay is required to make an accurate diagnosis of JEV genotype. In this study, we designed common and differential primer sets for JEV G1 and G3 to detect the JEV envelope (E) gene. The specific primer sets for JEV G1 and G3 specifically amplified the target gene. The detection limits of the three primer sets were 101.0, 102.0, and 102.0 TCID50/ reaction, respectively. No cross-reactivity was detected with non-JEV reference viruses. The multiplex reverse transcription-polymerase chain reaction (RT-PCR) assay specifically differentiated JEV G1 from G3. Thus, a one-step multiplex RT-PCR assay was established to rapidly and differentially detect JEV. This assay will be useful for confirming JEV infections in animals and checking the JEV genotype in veterinary biological products.
Go to : 
REFERENCES
1). Han N, Adams J, Chen P, Guo ZY, Zhong XF, Fang W, et al. Comparison of genotypes I and III in Japanese encephalitis virus reveals distinct differences in their genetic and host diversity. J Virol. 2014; 88:11469–79.

3). Aubry M, Finke J, Teissier A, Roche C, Broult J, Paulous S, et al. Seroprevalence of arboviruses among blood donors in French Polynesia, 2011–2013. Int J Infect Dis. 2015; 41:11–2.

4). Ravanini P, Huhtamo E, Ilaria V, Crobu MG, Nicosia AM, Servino L, et al. Japanese encephalitis virus RNA detected in Culex pipiens mosquitoes in Italy. Euro Surveill. 2012; 17:pii: 20221.

5). Gulati BR, Singha H, Singh BK, Virmani N, Kumar S, Singh RK. Isolation and genetic characterization of Japanese encephalitis virus from equines in India. J Vet Sci. 2012; 13:111–8.

6). Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001; 75:4268–75.

7). Chen WR, Tesh RB, Rico-Hesse R. Genetic variation of Japanese encephalitis virus in nature. J Gen Virol. 1990; 71(Pt 12):2915–22.

8). Hasegawa H, Yoshida M, Fujita S, Kobayashi Y. Comparison of structural proteins among antigenically different Japanese encephalitis virus strains. Vaccine. 1994; 12:841–4.

9). Kang BK, Hwang JM, Moon H, Han SY, Kim JM, Yang DK, et al. Comparison of the antigenic relationship between Japanese encephalitis virus genotypes 1 and 3. Clin Exp Vaccine Res. 2016; 5:26–30.

10). Nah JJ, Yang DK, Kim HH, Song JY. The present and future of veterinary vaccines for Japanese encephalitis in Korea. Clin Exp Vaccine Res. 2015; 4:130–6.

11). Zeng Z, Liu Z, Wang W, Tang D, Liang H, Liu Z. Establishment and application of a multiplex PCR for rapid and simultaneous detection of six viruses in swine. J Virol Methods. 2014; 208:102–6.

12). Kim HC, Takhampunya R, Tippayachai B, Chong ST, Park JY, Kim MS, et al. Japanese encephalitis virus in culicine mosquitoes (Diptera: culicidae) of the republic of Korea, 2008–2010. Mil Med. 2015; 180:158–67.

13). Yang DK, Kim BH, Kweon CH, Kwon JH, Lim SI, Han HR. Molecular characterization of full-length genome of Japanese encephalitis virus (KV1899) isolated from pigs in Korea. J Vet Sci. 2004; 5:197–205.

14). Yun SM, Cho JE, Ju YR, Kim SY, Ryou J, Han MG, et al. Molecular epidemiology of Japanese encephalitis virus circulating in South Korea, 1983–2005. Virol J. 2010; 7:127.

15). Yang DK, Kweon CH, Kim BH, Lim SI, Kim SH, Kwon JH, et al. TaqMan reverse transcription polymerase chain reaction for the detection of Japanese encephalitis virus. J Vet Sci. 2004; 5:345–51.

16). Zell R. Global climate change and the emergence/re-emergence of infectious diseases. Int J Med Microbiol. 2004; 293(Suppl 37):16–26.

17). Zheng H, Shan T, Deng Y, Sun C, Yuan S, Yin Y, et al. Molecular characterization of Japanese encephalitis virus strains prevalent in Chinese swine herds. J Vet Sci. 2013; 14:27–36.

18). Konishi E, Kitai Y, Tabei Y, Nishimura K, Harada S. Natural Japanese encephalitis virus infection among humans in west and east Japan shows the need to continue a vaccination program. Vaccine. 2010; 28:2664–70.

19). Yang DK, Nah JJ, Kim HH, Song JY. Inactivated genotype 1 Japanese encephalitis vaccine for swine. Clin Exp Vaccine Res. 2014; 3:212–9.

20). Chen YY, Lin JW, Fan YC, Chiou SS. Detection and differentiation of genotype I and III Japanese encephalitis virus in mosquitoes by multiplex reverse transcriptase-polymerase chain reaction. Transbound Emerg Dis. 2014; 61:37–43.

21). Wittwer CT, Herrmann MG, Gundry CN, Elenitoba-Johnson KS. Real-time multiplex PCR assays. Methods. 2001; 25:430–42.

22). Das S, Pingle MR, Munoz-Jordan J, Rundell MS, Rondini S, Granger K, et al. Detection and serotyping of dengue virus in serum samples by multiplex reverse transcriptase PCR-ligase detection reaction assay. J Clin Microbiol. 2008; 46:3276–84.

23). Johnson BW, Russell BJ, Lanciotti RS. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2005; 43:4977–83.

24). Kim H, Cha GW, Jeong YE, Lee WG, Chang KS, Roh JY, et al. Detection of Japanese encephalitis virus genotype V in Culex orientalis and Culex pipiens (Diptera: Culicidae) in Korea. PLoS One. 2015; 10:e0116547.
Go to : 
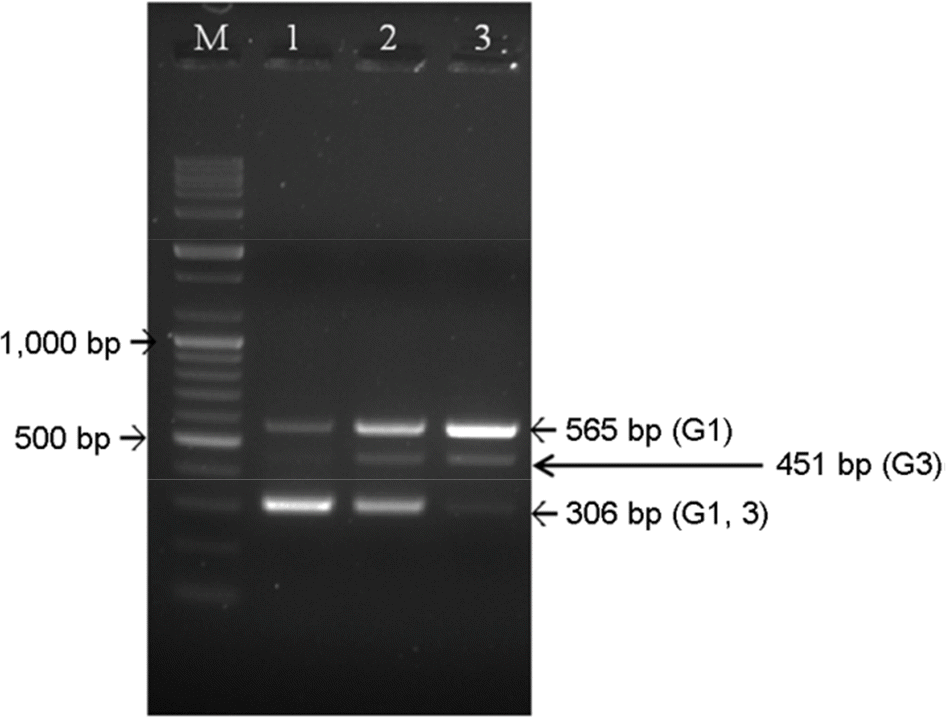 | Figure 2.Setting up annealing temperature to optimize temperature condition for detection of JEV genotypes. After conducting RT-PCR at three annealing temperature conditions (lane 1: 50°C, lane 2: 57°C, and lane 3: 62°C), the PCR products were visualized on the 1.8% agarose gel. JEV G3 cannot be amplified by RT-PCR of annealing temperature at 50°C and common parts of JEV G1 and G3 cannot be amplified distinctly at 62°C. Annealing temperature at 57°C turned to be reliable for detection of two JEV genotypes. |
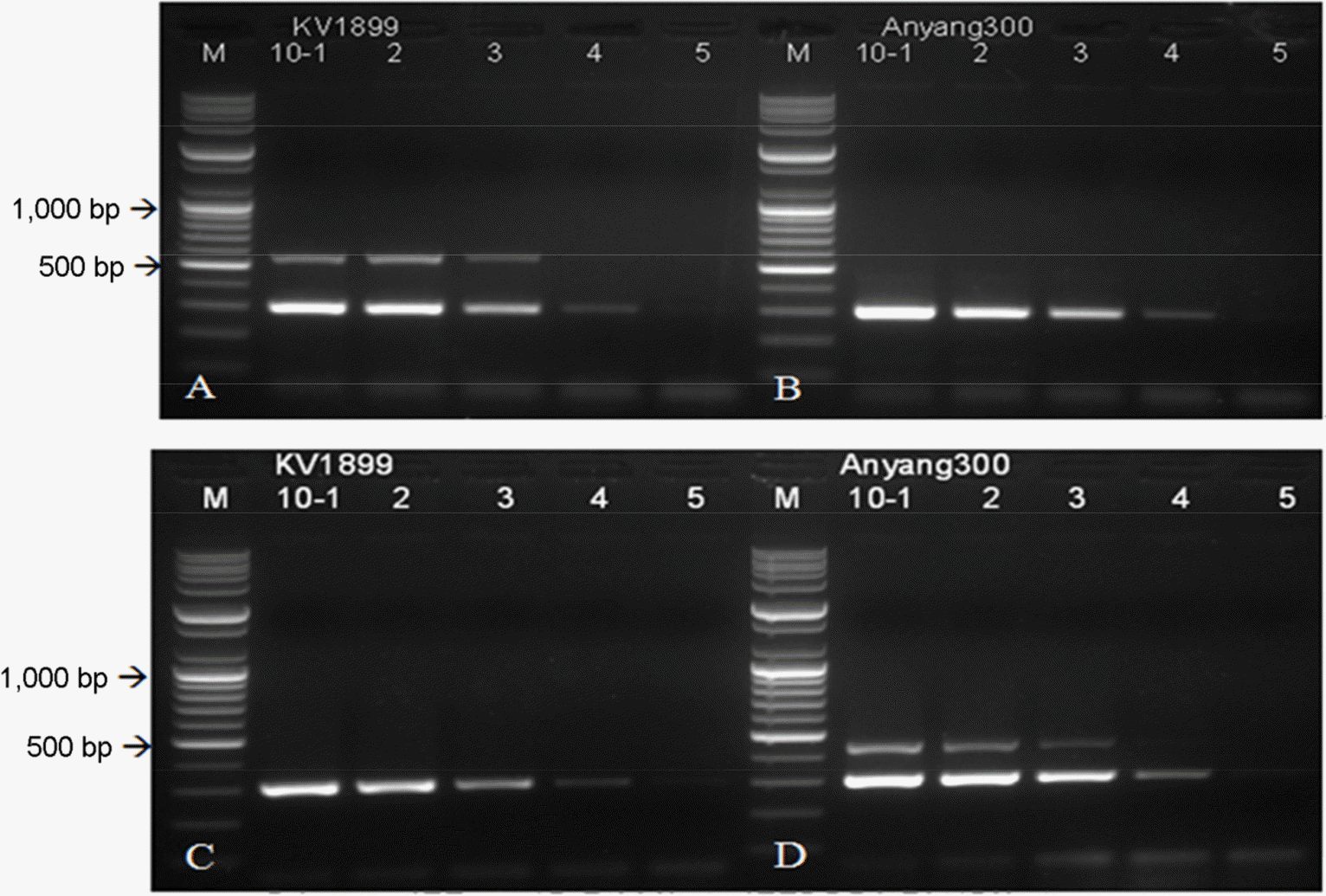 | Figure 3.Sensitivity of the specific primer sets for detection of genotypes of JEV. Differential RT-PCR between JEV G1 and G3 (A, B, C and D). Sensitivity of primer sets for common and genotypes was set up based on the annealing temperature. M; 100 bp DNA ladder, lane 1–5; 10-fold serial dilutions of extracted RNA of JEV (106.0 TCID50/ml). |
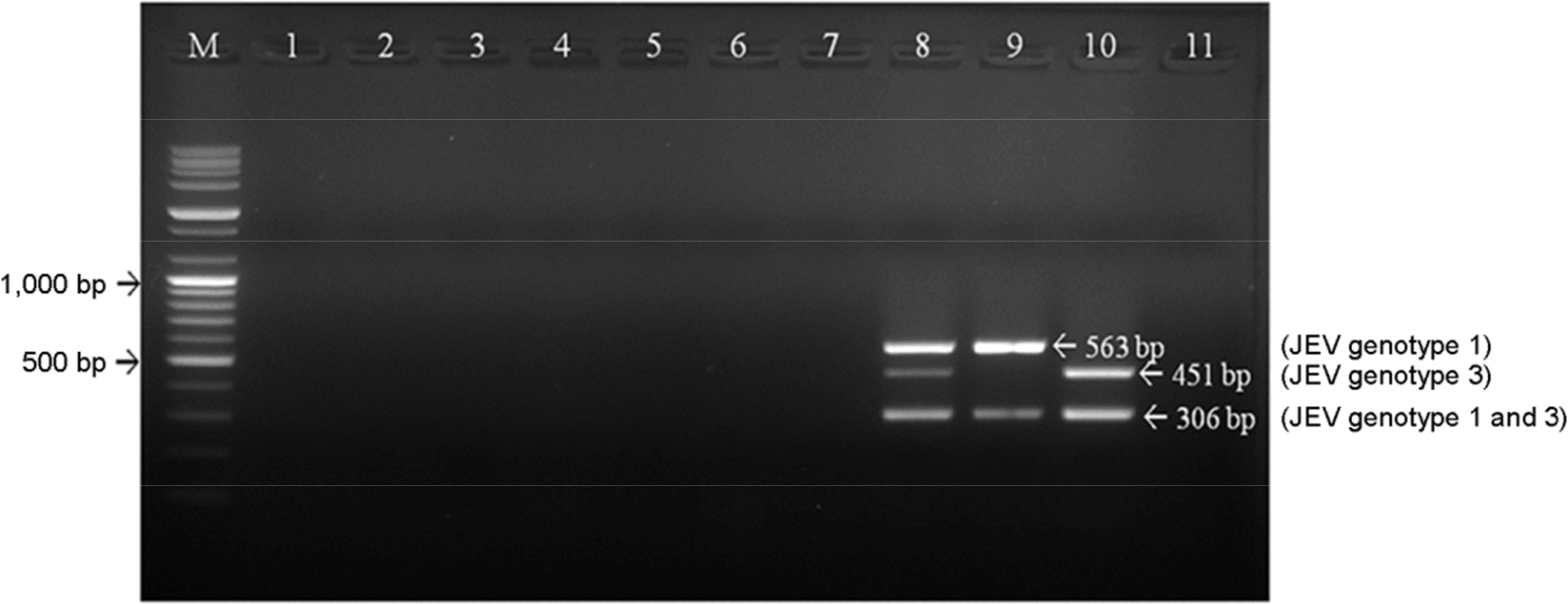 | Figure 4.Specificity of differential JEV RT-PCR using three kinds of primer sets. The RT-PCR detected only genotype of JEV and did not have positive signal for any other viral pathogens. The seven viruses showing titer of 105.0 TCID50/ml or over were used to extract RNA or DNA. M; 100 bp DNA ladder, lane 1; classical swine fever virus, lane 2; porcine parvovirus, lane 3; encephalomyocarditis virus, lane 4; Aujezsky's disease virus, lane 5; transmissible gastroenteritis virus, lane 6; porcine epidemic diarrhea virus, lane 7; porcine reproductive and respiratory syndrome virus, lane 8; JEV G1 and 3, lane 9; JEV G1, lane 10; JEV G3, lane 11; distilled water (negative control). |
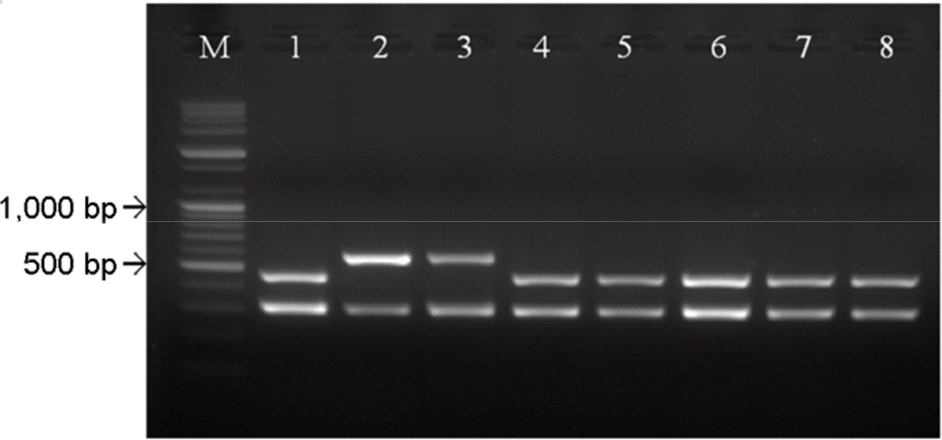 | Figure 5.Application of multiplex RT-PCR to commercial JE vaccines. Five JE vaccines and one Korean field strain (K95) showed positive reactions in the multiplex RT-PCR kit. M; 100 bp DNA ladder, lane 1; Anyang 300 strain, lane 2; KV1899 strain, lane 3; K95 strain, lane 4; Greencross® Porcine JE, lane 5; Daesung JE pig vac, lane 6; Suishot® JE, lane 7; Provac® JE, lane 8; Himmvac® JE. |
Table 1.
The design of the three JEV specific primer sets for differential detection of JEV in multiplex RT-PCR
| Name of primer | Genotype | Oligonucleotide sequences (5′ – 3′) | Discrepancy rate (%) | Position in E gene |
|---|---|---|---|---|
| JEcomF | G1 | CCAACACTAGATGTCCGCATGA | ||
| G3 | CCAACATTGGACGTCCGCATGA | 3/22 (13.6) | 115~135 | |
| Primer | CCAACAY∗ TR∗∗ GAYGTCCGCATGA | |||
| JEcomR | G1 | CATCAAGTACAAGGTTGGC | ||
| G3 | CATCAAATACGAAGTTGGC | 2/19 (10.5) | 402~420 | |
| Primer | CATCAAYTACYAYGTTGGC | |||
| JEG1F | G1 | GCGTCTCAAGCAGCAAAGTTTACT | ||
| G3 | GCGTCCCAGGCGGCAAAGTTTACA | 4/24 (16.7) | 481~504 | |
| Primer | GCGTCTCAAGCAGCAAAGTTTACT | |||
| JEG1R | G1 | TGTCTCAGTCGCGAGTTTAAACGAC | ||
| G3 | CGTCTCCGTTGCGAGCCTCAATGAC | 7/25 (28.0) | 1017~1043 | |
| Primer | TGTCTCAGTCGCGAGTTTAAACGAC | |||
| JEG3F | G1 | CCGATTGTCTCAGTCGCGAGTT | ||
| G3 | CCGATCGTCTCCGTTGCGAGCC | 5/22 (22.7) | 1012~1033 | |
| Primer | CCGATCGTCTCCGTTGCGAGCC | |||
| JEG3R | G1 | CCTGGCTTTTCTGGCCACGG | ||
| G3 | TTTGGCCTTCTTAGCCACAG | 7/20 (35.0) | 1443~1462 | |
| Primer | TTTGGCCTTCTTAGCCACAG |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download