Abstract
An increasing prevalence of infections caused by multidrug-resistant (MDR) Pseudomonas aeruginosa (P. aeruginosa) causes a serious therapeutic problem in clinical setting. This study investigated the antimicrobial susceptibility, resistance mechanisms against aminoglycosides, and molecular epidemiology of 76 blood isolates of P. aeruginosa from two Korean hospitals. Thirty-four isolates were susceptible to all 13 antimicrobial agents tested, whereas 28 isolates showed a MDR or extensively drug-resistant phenotype. There was a significant difference in resistance rates of P. aeruginosa isolates against aztreonam, piperacillin-tazobactam, imipenem, meropenem, ciprofloxacin, and norfloxacin between two hospitals. Genes for aminoglycoside-modifying enzymes (AMEs), including aphA6 (n = 14), aadB (n = 11), aacA4 (n = 8), and aphA1 (n = 1), and 16S rRNA methylase armA (n = 6) were detected in 26 P. aeruginosa isolates resistant to aminoglycosides. There was no significant difference in carriage of genes for AME and 16S rRNA methylase between two hospitals, but aacA4 and aphA1 were specifically detected in P. aeruginosa isolates from one hospital. Seventy-six P. aeruginosa isolates were classified into 55 pulsotypes at similarity value of 0.85, and 31 and 24 pulsotypes were specifically detected in each hospital. This study demonstrates that differences in antimicrobial susceptibility of P. aeruginosa isolates between two hospitals are possibly due to the presence of diverse clones specific in each hospital.
Go to : 
REFERENCES
1). Hidron AI, Edwards JR, Patel J, Horan TC, Sievert DM, Pollock DA, et al. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008; 29:996–1011.
2). Jones RN, Stilwell MG, Rhomberg PR, Sader HS. Antipseudomonal activity of piperacillin/tazobactam: more than a decade of experience from the SENTRY Antimicrobial Surveillance Program (1997–2007). Diagn Microbiol Infect Dis. 2009; 65:331–4.

3). Zhanel GG, DeCorby M, Adam H, Mulvey MR, McCracken M, Lagace-Wiens P, et al. Prevalence of antimicrobial-resistant pathogens in Canadian hospitals: results of the Canadian Ward Surveillance Study (CANWARD 2008). Antimicrob Agents Chemother. 2010; 54:4684–93.

4). Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24, 179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004; 39:309–17.
6). Mahar P, Padiglione AA, Cleland H, Paul E, Hinrichs M, Wasiak J. Pseudomonas aeruginosa bacteraemia in burns patients: Risk factors and outcomes. Burns. 2010; 36:1228–33.
7). Lambert ML, Suetens C, Savey A, Palomar M, Hiesmayr M, Morales I, et al. Clinical outcomes of health-care-associated infections and antimicrobial resistance in patients admitted to European intensive-care units: a cohort study. Lancet Infect Dis. 2011; 11:30–8.

8). Magnet S, Blanchard JS. Molecular insights into aminoglycoside action and resistance. Chem Rev. 2005; 105:477.

9). Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol. 2007; 5:939–51.
10). Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among strains of pan-European Acinetobacter baumannii clones. J Med Microbiol. 2004; 53:1233–40.
11). Lee H, Yong D, Yum JH, Roh KH, Lee K, Yamane K, et al. Dissemination of 16S rRNA methylase-mediated highly amikacin-resistant isolates of Klebsiella pneumoniae and Acinetobacter baumannii in Korea. Diagn Microbiol Infect Dis. 2006; 56:305–12.
12). Yokoyama K, Doi Y, Yamane K, Kurokawa H, Shibata N, Shibayama K, et al. Acquisition of 16S rRNA methylase gene in Pseudomonas aeruginosa. Lancet. 2003; 362:1888–93.
13). Cho YJ, Moon DC, Jin JS, Choi CH, Lee YC, Lee JC. Genetic basis of resistance to aminoglycosides in Acinetobacter spp. and spread of armA in Acinetobacter baumannii sequence group 1 in Korean hospitals. Diagn Microbiol Infect Dis. 2009; 64:185–90.
14). Gurung M, Moon DC, Tamang MD, Kim J, Lee YC, Seol SY, et al. Emergence of 16S rRNA methylase gene armA and cocarriage of bla (IMP-1) in Pseudomonas aeruginosa isolates from South Korea. Diagn Microbiol Infect Dis. 2010; 68:468–70.
15). Kim CH, Kang HY, Kim BR, Jeon H, Lee YC, Lee SH, et al. Mutational inactivation of OprD in carbapenem-resistant Pseudomonas aeruginosa isolates from Korean hospitals. J Microbiol. 2016; 54:44–9.
16). Jin JS, Kwon KT, Moon DC, Lee JC. Emergence of 16S rRNA methylase rmtA in colistin-only-sensitive Pseudomonas aeruginosa in South Korea. Int J Antimicrob Agents. 2009; 33:490–1.
17). Clinical Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fifth Informational Supplement M100-S25. Wayne, PA, USA: CLSI;. 2015.
18). Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18:268–81.

19). Yan JJ, Wu JJ, Ko WC, Tsai SH, Chuang CL, Wu HM, et al. Plasmid-mediated 16S rRNA methylases conferring high-level aminoglycoside resistance in Escherichia coli and Klebsiella pneumoniae isolates from two Taiwanese hospitals. J Antimicrob Chemother. 2004; 54:1007–12.
20). Wachino J, Yamane K, Shibayama K, Kurokawa H, Shibata N, Suzuki S, et al. Novel plasmid-mediated 16S rRNA methylase, RmtC, found in a proteus mirabilis isolate demonstrating extraordinary high-level resistance against various aminoglycosides. Antimicrob Agents Chemother. 2006; 50:178–84.
21). Fritsche TR, Castanheira M, Miller GH, Jones RN, Armstrong ES. Detection of methyltransferases conferring high-level resistance to aminoglycosides in enterobacteriaceae from Europe, North America, and Latin America. Antimicrob Agents Chemother. 2008; 52:1843–5.

22). Ploy MC, Giamarellou H, Bourlioux P, Courvalin P, Lambert T. Detection of aac(6′)-I genes in amikacin-resistant Acinetobacter spp. by PCR. Antimicrob Agents Chemother. 1994; 38:2925–8.
23). Vila J, Ruiz J, Navia M, Becerril B, Garcia I, Perea S, et al. Spread of amikacin resistance in Acinetobacter baumannii strains isolated in Spain due to an epidemic strain. J Clin Microbiol. 1999; 37:758–61.
24). Noppe-Leclercq I, Wallet F, Haentjens S, Courcol R, Simonet M. PCR detection of aminoglycoside resistance genes: a rapid molecular typing method for Acinetobacter baumannii. Res Microbiol. 1999; 150:317–22.

25). Lee YC, Ahn BJ, Jin JS, Kim JU, Lee SH, Song DY, et al. Molecular characterization of Pseudomonas aeruginosa isolates resistant to all antimicrobial agents, but susceptible to colistin, in Daegu, Korea. J Microbiol. 2007; 45:358–63.
26). Lee J, Oh CE, Choi EH, Lee HJ. The impact of the increased use of piperacillin/tazobactam on the selection of antibiotic resistance among invasive Escherichia coli and Klebsiella pneumoniae isolates. Int J Infect Dis. 2013; 17:e638–43.
Go to : 
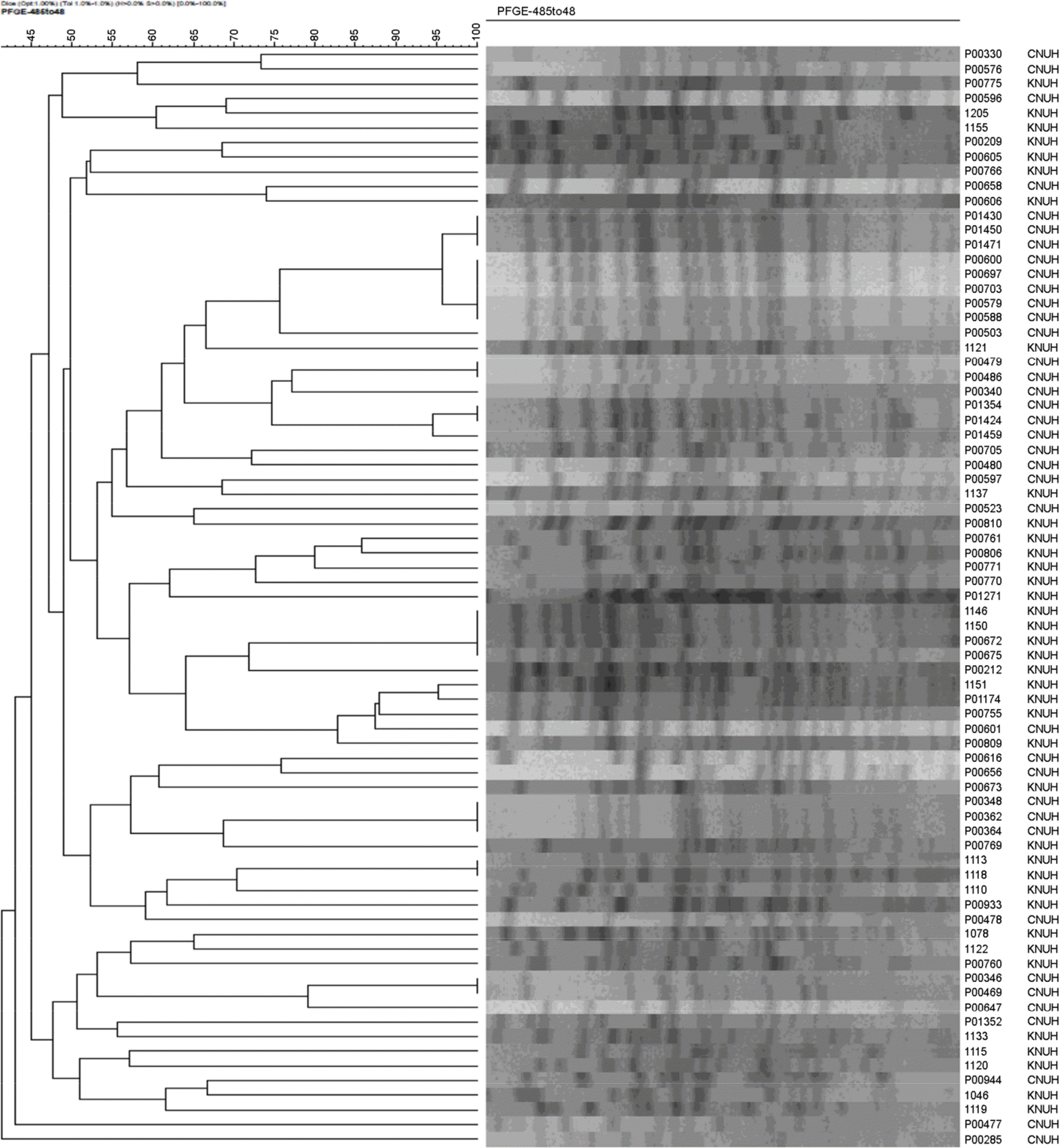 | Figure 1.Pulsed-field gel electrophoresis analysis of 76 P. aeruginosa isolates from two Korean hospitals. Dendrogram showing a clustering of Spe Igenerated PFGE patterns from P. aeruginosa isolates. Abbreviations: KNUH, Hospital A; CNUH, hospital B. |
Table 1.
PCR primers used in this study
Table 2.
Antimicrobial susceptibility of P. aeruginosa isolates from two Korean hospitals
Table 3.
Antimicrobial susceptibility profiles that fit MDR, XDR, and PDR for 76 P. aeruginosa isolates
| Antimicrobial susceptibility profilesa | No. of isolates (%) | ||
|---|---|---|---|
| Total (n = 76) | Hospital A (n = 39) | Hospital B (n = 37) | |
| Susceptible to antimicrobial agents | 34 (44.7) | 20 (51.3) | 14 (37.8) |
| Resistant to 1 or 2 antimicrobial categories | 14 (18.4) | 9 (23.1) | 5 (13.5) |
| Multidrug-resistant | 14 (18.4) | 6 (15.4) | 8 (21.6) |
| Extensively drug-resistant | 14 (18.4) | 4 (10.3) | 10 (27.0) |
| Pandrug-resistant | 0 | 0 | 0 |
a Multidrug-resistant (MDR), the isolate is non-susceptible to at least 1 agent in ≥ 3 antimicrobial classes; extensively drug-resistant (XDR), the isolate is non-susceptible to at least 1 agent in all but 2 or fewer antimicrobial classes; pandrug-resistant (PDR), nonsusceptibility to all agents in all antimicrobial classes for each bacterium.
Table 4.
Detection of resistant determinants for aminoglycosides in P. aeruginosa isolates from two Korean hospitals
| Resistant pattern of aminoglycosidesa | No. of isolates | Genes | No. of isolates | |
|---|---|---|---|---|
| Hospital A | Hospital B | |||
| aadB, armA | 1 | 4 | ||
| aphA6, aadB | – | 4 | ||
| Amk, Gem, Tob, Ntl | 13 | aacA4, aphA6, aadB | 2 | – |
| aacA4, aphA6, armA | 1 | – | ||
| Not detected | – | 1 | ||
| Gem, Tob, Ntl | 11 | aacA4 | 4 | – |
| aphA6 | – | 7 | ||
| Amk, Gem | 1 | Not detected | 1 | – |
| Gem, Ntl | 1 | aphA1 | 1 | – |
| Ntl, Tob | 1 | aacA4 | 1 | – |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download