Abstract
Species identification is an important item to characterize unidentified bacterial pathogens in developing and managing bacterial resources. In this study, unidentified pathogens based on the results of an automated identification system were identified using matrix assisted laser desorption ionization-time of flight mass spectrometry (MALD-TOF MS) and 16S rRNA gene analysis for development of national resources in the National Culture Collection for Pathogens (NCCP) in Korea. A total of 437 unidentified strains from branch banks of the NCCP were collected, and 16S rRNA and dnaJ gene sequencing, as well as MALDI-TOF MS analysis were performed to identify bacterial species. The mass spectra extracted were analyzed. Twelve strains exhibiting less than 98.65% similarity in 16S rRNA gene were selected as the primary candidates for novel species, and 21 strains exhibiting 98.65~99.0% similarity in 16S rRNA gene were selected as possible candidates for novel species. Among them, strain 32, belonging to Dermabacter sp., was finally selected as a possible strain representing a novel species and 14 unidentified bacterial strains using automated phenotypic identification system were newly registered at NCCP. The present study showed that unidentified pathogens using the automated phenotypic identification system were efficiently identified using the combination of MALDI-TOF MS and 16S rRNA gene analysis, and developed to the national resources in NCCP.
Go to : 
REFERENCES
1). Yu WS, Lee KM, Hyung KJ. Resource development of unidentified Human Pathogens using automated identification systems. Public Health Weekly Report. 2015; 8:990–7.
2). Funke G, Monnet D, deBernardis C, von Graevenitz A, Freney J. Evaluation of the VITEK 2 system for rapid identification of medically relevant gram-negative rods. J Clin Microbiol. 1998; 36:1948–52.

3). Benagli C, Demarta A, Caminada A, Ziegler D, Petrini O, Tonolla M. A rapid MALDI-TOF MS identification database at genospecies level for clinical and environmental Aeromonas strains. PLoS One. 2012; 7:e48441.

4). Christner M, Trusch M, Rohde H, Kwiatkowski M, Schluter H, Wolters M, et al. Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-Toxigenic Escherichia coli. PLoS One. 2014; 9:e101924.
5). Josten M, Reif M, Szekat C, Al-Sabti N, Roemer T, Sparbier K, et al. Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J Clin Microbiol. 2013; 51:1809–17.
6). Khot PD, Fisher MA. Novel approach for differentiating Shigell a species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013; 51:3711–6.
7). Petti CA, Bosshard PP, Brandt ME, Clarridge JE, Feldblyum TV, Foxall P, et al. Interpretative criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. MM18-A. 2008; 28:1–7.
8). Rozhon W, Khan M, Poppenberger B. The natural antibiotic resistances of the Enterobacteriaceae Rahnella and Ewingell a. In: Pana M, editor. Antibiotic resistant bacteria a continuous challenge in the new millennium. InTech;. 2012. 77–104.

9). Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62:716–21.

10). Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013; 30:2725–9.

11). Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004; 5:150–63.

12). Jukes TH, Cantor CR. Evolution of protein molecules. Munro HN, editor. Mammalian protein metabolism. New York: Academic Press;1960. p. 21–132.

13). Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987; 4:406–25.
14). Haigh J, Degun A, Eydmann M, Millar M, Wilks M. Improved performance of bacterium and yeast identification by a commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry system in the clinical microbiology laboratory. J Clin Microbiol. 2011; 49:3441.

15). Schulthess B, Bloemberg GV, Zbinden R, Bottger EC, Hombach M. Evaluation of the Bruker MALDI Biotyper for identification of Gram-positive rods: development of a diagnostic algorithm for the clinical laboratory. J Clin Microbiol. 2014; 52:1089–97.

16). Schulthess B, Brodner K, Bloemberg GV, Zbinden R, Bottger EC, Hombach M. Identification of Gram-positive cocci by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry: comparison of different preparation methods and implementation of a practical algorithm for routine diagnostics. J Clin Microbiol. 2013; 51:1834–40.

17). Yang C, He Z, Yu W. Comparison of public peak detection algorithms for MALDI mass spectrometry data analysis. BMC Bioinformatics. 2009; 10:4.

18). Stanford, TE, Bagley CJ, Solomon. Informed baseline subtraction of proteomic mass spectrometry data aided by a novel sliding window algorithm. 2016. arXiv: 1603.07082 [q-bio.QM].
19). Kim M, Oh HS, Park SC, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014; 64:346–51.

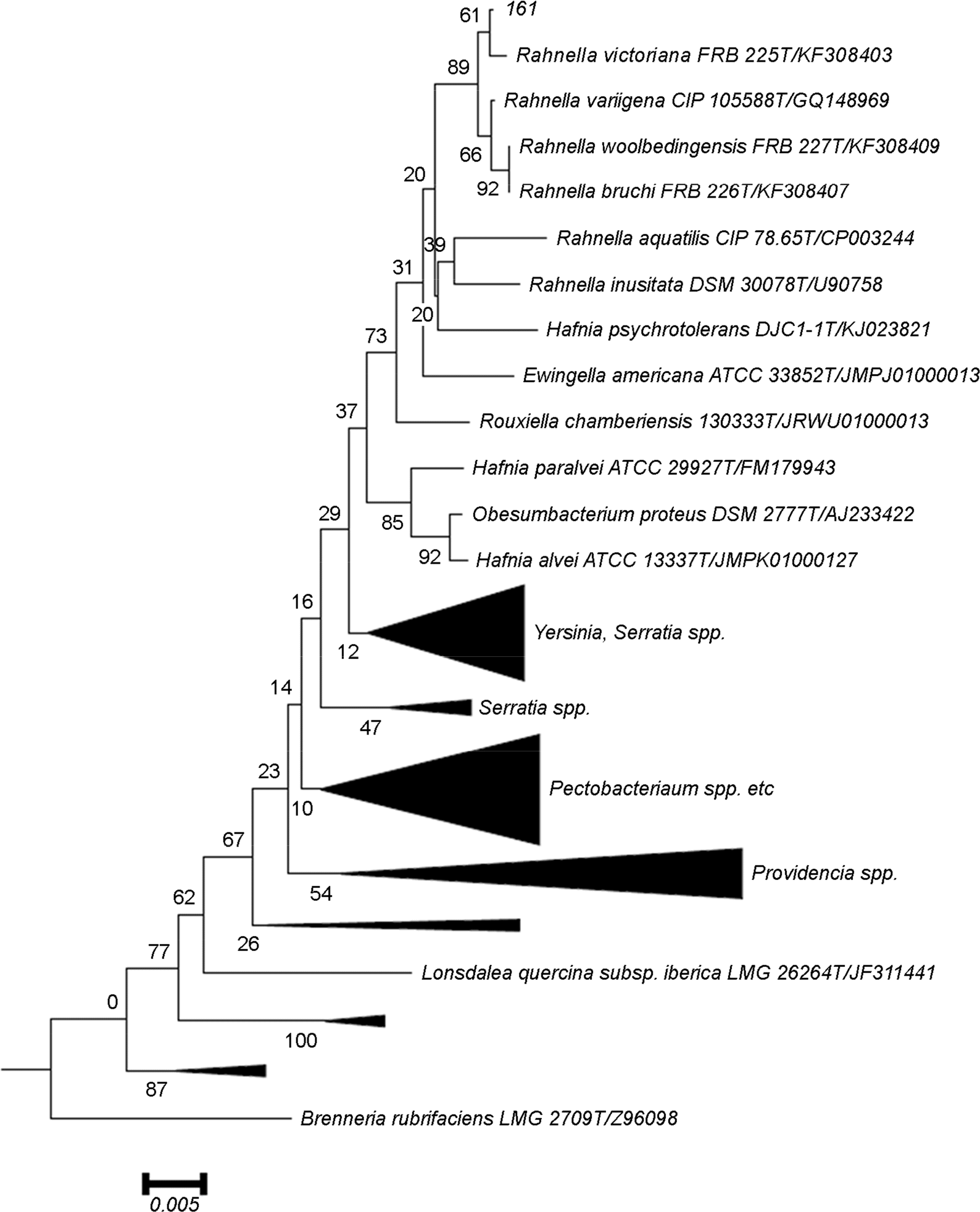
20). Brady C, Hunter G, Kirk S, Arnold D, Denman S. Rahnella victoriana sp. nov., Rahnella bruchi sp. nov., Rahnella woolbedingensis sp. nov., classification of Rahnella genomospecies 2 and 3 as Rahnella variigena sp. nov. and Rahnella inusitata sp. nov., respectively and emended description of the genus Rahnella. Syst Appl Microbiol. 2014; 37:545–52.
21). Ezaki T, Hashimoto Y, Yabuuchi E. Fluorometric deoxy-ribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Evol Microbiol. 1989; 39:224–9.

22). Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Evol Microbiol. 1987; 37:463–4.

23). Tindall BJ, Rossello-Mora R, Busse HJ, Ludwig W, Kampfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol. 2010; 60:249–66.

24). Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, et al. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect. 2010; 16:998–1004.

25). Jamal W, Albert MJ, Rotimi VO. Real-time comparative evaluation of bioMerieux VITEK MS versus Bruker Microflex MS, two matrix-assisted laser desorption-ionization time-of-flight mass spectrometry systems, for identification of clinically significant bacteria. BMC Microbiol. 2014; 14:289.

26). Schaumann R, Knoop N, Genzel GH, Losensky K, Rosenkranz C, Stingu CS, et al. A step towards the discrimination of beta-lactamase-producing clinical isolates of Enterobacteriaceae and Pseudomonas aeruginosa by MALDI-TOF mass spectrometry. Med Sci Monit. 2012; 18:MT71–7.

27). Hart PJ, Wey E, McHugh TD, Balakrishnan I, Belgacem O. A method for the detection of antibiotic resistance markers in clinical strains of Escherichia coli using MALDI mass spectrometry. J Microbiol Methods. 2015; 111:1–8.
Go to : 
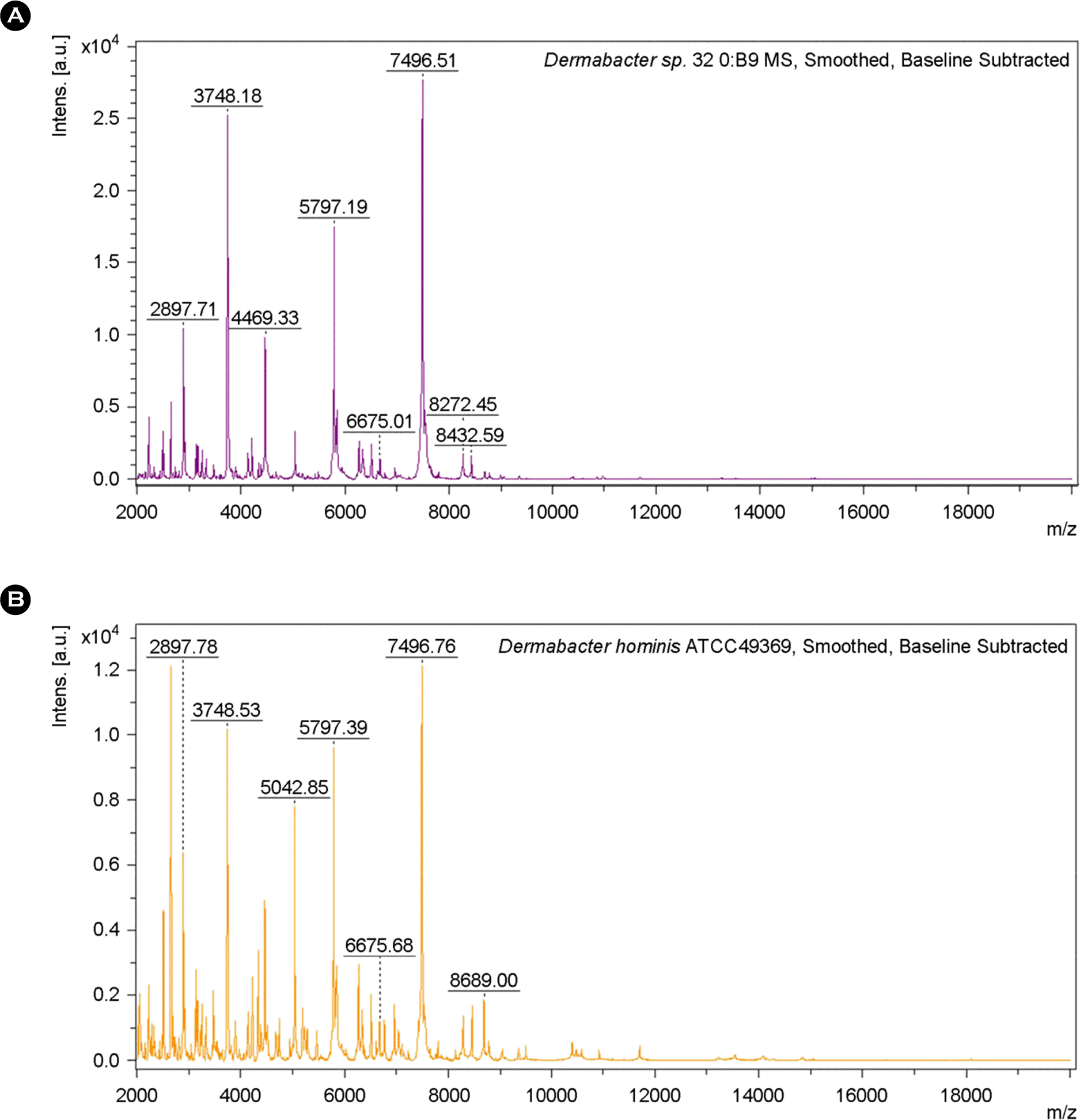 | Figure 5.Schematic representation of single major spectrometric profile from strain 32 (a) and Dermabacter hominis ATCC49369 (b). |
Table 1.
Clinical information of newly-registered bacterial resources
Table 2.
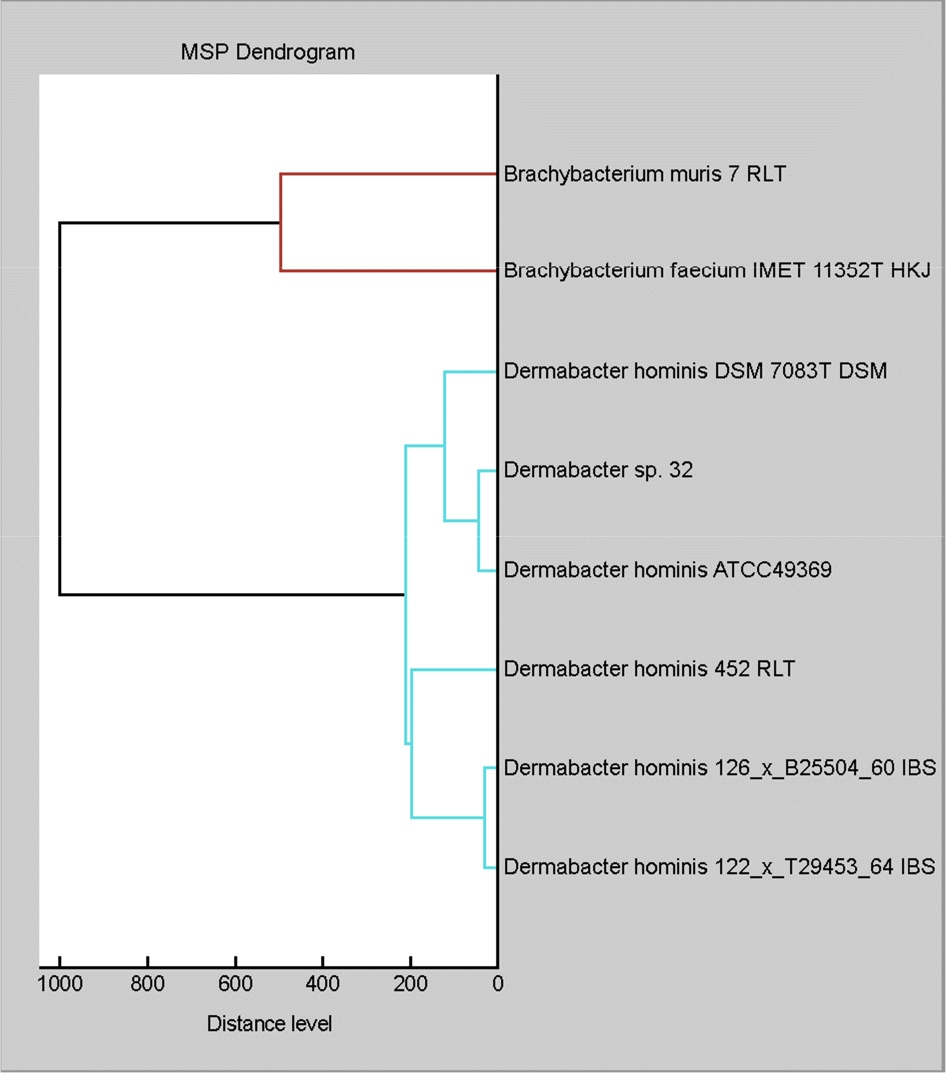
Identification results of strain 32 and Dermabacter hominis ATCC49369 using MALDI-TOF MS and 16S rRNA gen
Table 3.
Biochemical characteristics of strain 32




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download