Abstract
Propionibacterium acnes, a gram-positive, anaerobic, and aerotolerant bacterium that is found frequently in the skin as part of the human microbiome causes inflammatory acne, shoulder infection, and the contamination of medical devices. The study goals were the antibiotic resistant and molecular epidemiological characterization of the P. acnes isolates in Korea. A total of 22 P. acnes isolates originated from diverse patients were obtained from three National Culture Collections for Pathogens in South Korea. The hemolytic properties and minimum inhibitory concentrations (MIC) of five antibiotics (tetracycline, doxycycline, clindamycin, erythromycin, and minocycline) were determined. Only one isolate showed high MIC values and resistance to all five antibiotics. Genotypic characterization was achieved by multilocus sequence typing (MLST) for eight loci (aroE, guaA, tly, camp2, atpD, gmk, lepA, and sodA) and repetitive-sequence-based PCR (rep-PCR) analysis using the DiversiLab kit. MLST revealed four phylogroups that were type IA1 (27.3%), type IA2 (18.2%), type IB (13.6%), and type II (40.9%). Rep-PCR results demonstrated three clusters that were cluster I (39.1%), cluster II (45.5%), and cluster III (13.6%). The isolates of cluster I were part of phylogroup type IA (both IA1 and IA2), and the isolates of cluster II belonged to phylogroup type II. All isolates of phylogroup type IB were hemolytic and belonged to cluster III. The results of rep-PCR clustering analysis showed a good correlation with those of MLST phylogroups, suggesting that rep-PCR could be an alternative method to track P. acnes subtype lineages.
Go to : 
REFERENCES
1). Cogen AL, Nizet V, Gallo R. Skin microbiota: a source of disease or defence? Br J Dermatol. 2008; 158:442–55.

2). Fitz-Gibbon S, Tomida S, Chiu BH, Nguyen L, Du C, Liu M, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013; 133:2152–60.
3). Eishi Y, Suga M, Ishige I, Kobayashi D, Yamada T, Takemura T, et al. Quantitative analysis of mycobacterial and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J Clin Microbiol. 2002; 40:198–204.

4). Fassi Fehri L, Mak TN, Laube B, Brinkmann V, Ogilvie LA, Mollenkopf H, et al. Prevalence of Propionibacterium acnes in diseased prostates and its inflammatory and transforming activity on prostate epithelial cells. Int J Med Microbiol. 2011; 301:69–78.
5). Perry A, Lambert P. Propionibacterium acnes: infection beyond the skin. Expert Rev Anti Infect Ther. 2011; 9:1149–56.
6). Ishige I, Eishi Y, Takemura T, Kobayashi I, Nakata K, Tanaka I, et al. Propionibacterium acnes is the most common bacterium commensal in peripheral lung tissue and mediastinal lymph nodes from subjects without sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 2005; 22:33–42.
7). Mollerup S, Friis-Nielsen J, Vinner L, Hansen TA, Richter SR, Fridholm H, et al. Propionibacterium acnes: Disease-causing agent or common contaminant? Detection in diverse patient samples by next-generation sequencing. J Clin Microbiol. 2016; 54:980–7.
8). McDowell A, Gao A, Barnard E, Fink C, Murray PI, Dowson CG, et al. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology. 2011; 157:1990–2003.
9). McDowell A, Barnard E, Nagy I, Gao A, Tomida S, Li H, et al. An expanded multilocus sequence typing scheme for Propionibacterium acnes: investigation of ‘pathogenic’, ‘commensal’ and antibiotic resistant strains. PLoS One. 2012; 7:e41480.
10). Kilian M, Scholz CF, Lomholt HB. Multilocus sequence typing and phylogenetic analysis of Propionibacterium acnes. J Clin Microbiol. 2012; 50:1158–65.
11). Johnson T, Kang D, Barnard E, Li H. Strain-level differences in porphyrin production and regulation in Propionibacterium acnes elucidate disease associations. mSphere. 2016; 1:e00023–15.

12). Song M, Seo SH, Ko HC, Oh CK, Kwon KS, Chang CL, et al. Antibiotic susceptibility of Propionibacterium acnes isolated from acne vulgaris in Korea. J Dermatol. 2011; 38:667–73.
13). Ki HG, Yun SJ, Lee JB, Kim SJ, Lee SC, Won YH. Microorganisms isolated from acne and their antibiotic susceptibility. Kor J Dermatol. 2005; 43:871–5.
14). McDowell A, Perry AL, Lambert PA, Patrick S. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol. 2008; 57:218–24.
15). Schafer F, Fich F, Lam M, Gárate C, Wozniak A, Garcia P. Antimicrobial susceptibility and genetic characteristics of Propionibacterium acnes isolated from patients with acne. Int J Dermatol. 2013; 52:418–25.
16). McDowell A, Nagy I, Magyari M, Barnard E, Patrick S. The opportunistic pathogen Propionibacterium acnes: insights into typing, human disease, clonal diversification and CAMP factor evolution. PLoS One. 2013; 8:e70897.
17). Nodzo SR, Hohman DW, Crane JK, Duquin TR. Hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection. Am J Orthop. 2014; 43:E93–7.
18). Davidsson S, Söderquist B, Elgh F, Olsson J, Andrén O, Unemo M, et al. Multilocus sequence typing and repetitive-sequence-based PCR (DiversiLab) for molecular epidemiological characterization of Propionibacterium acnes isolates of heterogeneous origin. Anaerobe. 2012; 18:392–9.
19). Barnard E, Nagy I, Hunyadkurti J, Patrick S, McDowell A. Multiplex touchdown PCR for rapid typing of the opportunistic pathogen Propionibacterium acnes. J Clin Microbiol. 2015; 53:1149–55.
Go to : 
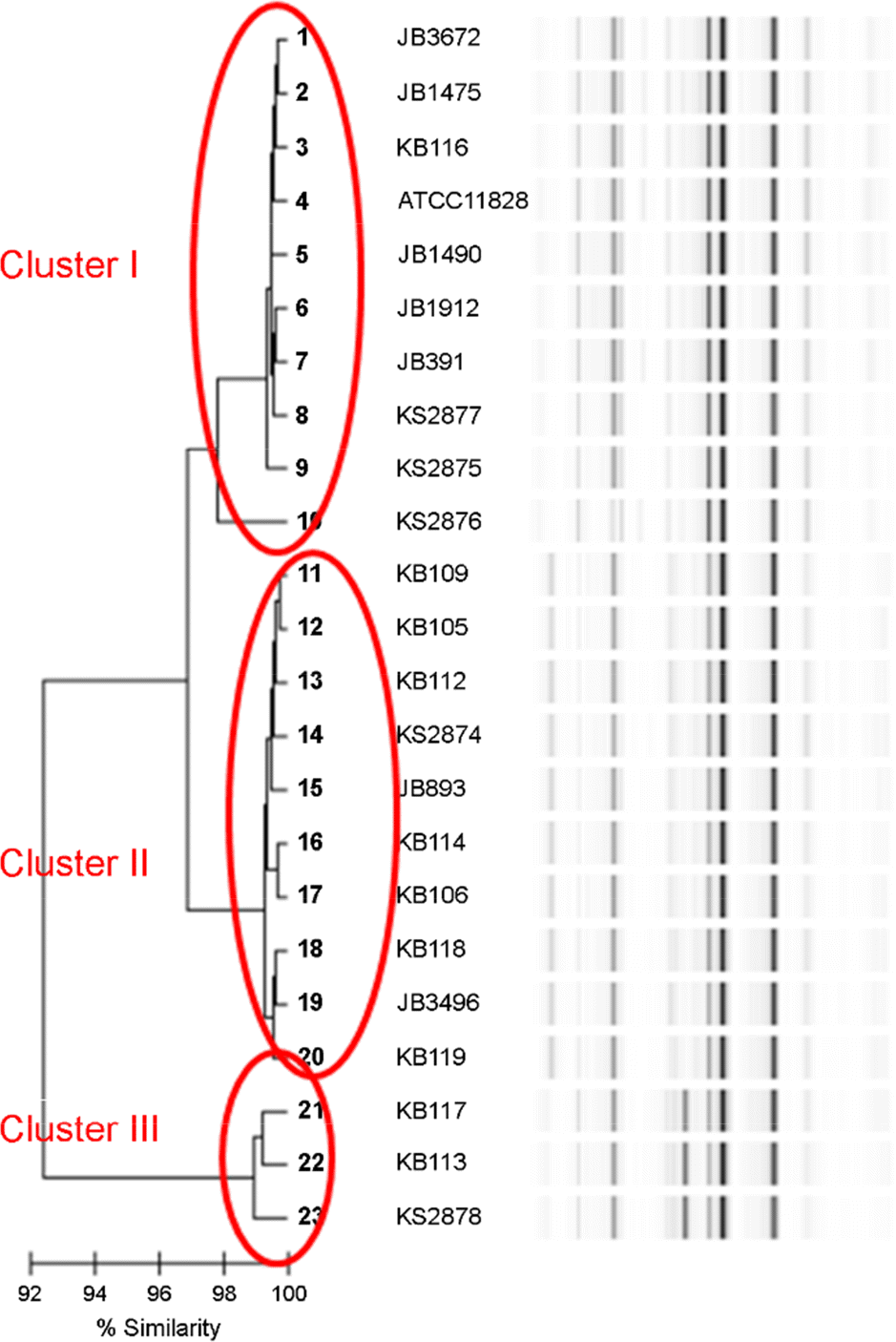 | Figure 1.Dendrogram and three similarity clusters generated from repetitive-sequence-based PCR results for 22 clinical isolates of P. acnes and one type strain, P. acnes ATCC 11828. |
Table 1.
Minimum inhibitory concentrations (µg/ml) of five antimicrobial agents using 22 clinical isolates and one standard strain of P. acnes
| P. acnes isolate | Tetracycline (RB1: 2.0) | Doxycycline (RB: 1.0) | Clindamycin (RB: 4.0) | Erythromycin (RB: 0.5) | Minocycline (RB: 1.0) |
|---|---|---|---|---|---|
| KB105 | 0.064 | 0.125 | ≤ 0.016 | ≤ 0.016 | 0.047 |
| KB106 | 0.25 | 0.19 | ≤ 0.016 | ≤ 0.016 | ≤ 0.016 |
| KB109 | 0.25 | 0.38 | 0.023 | ≤ 0.016 | 0.094 |
| KB112 | 8 | 4 | ≥ 256 | ≥ 256 | 3 |
| KB113 | 0.125 | 0.125 | ≤ 0.016 | 0.023 | 0.047 |
| KB114 | 0.25 | 0.25 | ≤ 0.016 | 0.023 | 0.125 |
| KB116 | ≤ 0.016 | 0.19 | ≤ 0.016 | ≤ 0.016 | ≤ 0.016 |
| KB117 | 0.064 | 0.064 | 0.023 | ≤ 0.016 | 0.023 |
| KB118 | 0.064 | 0.032 | ≤ 0.016 | ≤ 0.016 | 0.125 |
| KB119 | 0.5 | 0.25 | ≤ 0.016 | ≤ 0.016 | 0.047 |
| JB391 | 0.125 | 0.047 | ≤ 0.016 | ≤ 0.016 | 0.023 |
| JB893 | 0.25 | 0.5 | 0.023 | ≤ 0.016 | 0.19 |
| JB1475 | 0.125 | 0.094 | ≤ 0.016 | ≤ 0.016 | 0.047 |
| JB1490 | 0.125 | 0.125 | ≤ 0.016 | ≤ 0.016 | 0.023 |
| JB1912 | 0.25 | 0.25 | 0.023 | ≤ 0.016 | 0.047 |
| JB3672 | 0.047 | 0.032 | ≤ 0.016 | ≤ 0.016 | ≤ 0.016 |
| JB3496 | 0.064 | 0.064 | ≤ 0.016 | ≤ 0.016 | 0.023 |
| KS2874 | 0.25 | 0.064 | ≤ 0.016 | ≤ 0.016 | ≤ 0.016 |
| KS2875 | 0.5 | 0.125 | 0.032 | ≤ 0.016 | 0.094 |
| KS2876 | 0.19 | 0.19 | 0.023 | ≤ 0.016 | 0.047 |
| KS2877 | 0.25 | 0.19 | 0.032 | ≤ 0.016 | 0.032 |
| KS2878 | 0.125 | 0.125 | 0.023 | ≤ 0.016 | ≤ 0.016 |
| ATCC 11828 | 0.19 | 0.125 | ≤ 0.016 | ≤ 0.016 | 0.047 |
Table 2.
Hemolysis, multilocus sequence typing (MLST), and repetitive-sequence-based PCR (rep-PCR) for 22 clinical isolates and 1 type strain of P. acnes
| Isolate | Hemolysis1 | MLST | Rep-PCR p | |||
|---|---|---|---|---|---|---|
| Allelic profile (aroE-guaA-tly-camp2-atpD- gmk-lepA-sodA) | ST | CC | Phylogroup | |||
| KB105 | - | 1-5-8-2-1-1-1-4 | 2 | CC2 | IA2 | Cluster I |
| KB106 | - | 1-5-8-7-1-1-1-5 | 22 | Singleton | IA2 | Cluster I |
| KB109 | + | 1-3-1-1-1-1-1-1 | 1 | CC1 | IA1 | Cluster I |
| KB112 | + | 1-3-22-2-1-1-1-1 | 115 | CC3 | IA1 | Cluster I |
| KB113 | + | 1-4-8-6-1-9-1-4 | 53 | CC5 | IB | Cluster III |
| KB114 | + | 1-5-8-7-1-1-1-5 | 22 | Singleton | IA2 | Cluster I |
| KB116 | - | 17-4-10-12-4-2-4-6 | 69 | CC72 | II | Cluster II |
| KB117 | + | 1-4-8-6-1-1-1-4 | 5 | CC5 | IB | Cluster III |
| KB118 | + | 1-3-1-1-1-1-1-1 | 1 | CC1 | IA1 | Cluster I |
| KB119 | + | 1-3-1-1-1-1-1-1 | 1 | CC1 | IA1 | Cluster I |
| JB391 | + | 17-4-10-12-4-2-4-6 | 69 | CC72 | II | Cluster II |
| JB893 | + | 1-3-1-1-1-1-1-1 | 1 | CC1 | IA1 | Cluster I |
| JB1475 | - | 17-4-10-12-4-2-4-6 | 69 | CC72 | II | Cluster II |
| JB1490 | - | 17-4-10-10-4-2-2-3 | 6 | CC6 | II | Cluster II |
| JB1912 | - | 17-4-10-10-4-2-2-3 | 6 | CC6 | II | Cluster II |
| JB3496 | + | 1-3-22-2-1-1-1-1 | 115 | CC3 | IA1 | Cluster I |
| JB3672 | - | 17-4-10-12-4-2-4-6 | 69 | CC72 | II | Cluster II |
| KS2874 | - | 1-5-8-2-1-1-1-4 | 2 | CC2 | IA2 | Cluster I |
| KS2875 | - | 17-4-10-10-4-2-2-3 | 6 | CC6 | II | Cluster II |
| KS2876 | - | 17-4-10-12-4-2-4-6 | 69 | CC72 | II | Cluster II |
| KS2877 | - | 17-4-10-10-9-2-2-3 | 25 | CC6 | II | Cluster II |
| KS2878 | + | 1-4-8-6-1-9-1-4 | 53 | CC5 | IB | Cluster III |
| ATCC 11828 | - | 17-4-10-12-4-2-4-6 | 69 | CC72 | II | Cluster I |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download