Abstract
Turnip yellow mosaic virus (TYMV) is a non-enveloped icosahedral virus composed of 20 kDa single coat proteins. In this study, we modified the TYMV coat protein (CP) ORF by inserting an oligonucleotide linker corresponding to T7, HSV, Tat, (Arg)9, or (RxR)4 peptide at the 5′-end of the CP ORF and examined its effect on replication, RNA packaging, and virion assembly. The results showed that the constructs containing (Arg)9 and (RxR)4 sequences were barely capable of replication. The TYMV constructs containing T7 and Tat peptide produced virions that co-migrated with wild-type virions. However, the insertion of T7 and Tat sequences impaired genomic RNA (gRNA) accumulation and packaging, respectively. When only the CP gene was expressed, CPs with (Arg)9 or (RxR)4 successfully produced virus-like particles whose mobility was comparable to that of wild type. In the case of CP having a HSV tag, the virion band was not detected, although a sufficient amount of CP was produced. This indicates that CP with the HSV tag failed to assemble into virions. Overall, the results suggest that TYMV replication, RNA packaging and virion assembly are strongly influenced by the insertion sequence.
Go to : 
REFERENCES
1). Dreher TW. Turnip yellow mosaic virus: transfer RNA mimicry, chloroplasts and a C-rich genome. Mol Plant Pathol. 2004; 5:367–75.

2). Canady MA, Larson SB, Day J, McPherson A. Crystal structure of turnip yellow mosaic virus. Nat Struct Biol. 1996; 3:771–81.

3). Steinmetz NF, Evans DJ. Utilization of plant viruses in biotechnology. Org Biomol Chem. 2007; 5:2891–902.
4). Steinmetz NF. Viral nanoparticles as platforms for next-generation therapeutics and imaging devices. Nanomedicine. 2010; 6:634–41.

5). Yildiz I, Shukla S, Steinmetz NF. Applications of viral nanoparticles in medicine. Curr Op Biotechnol. 2011; 22:901–8.

6). Brundel FM, Lewis JD, Desitto G, Steinmetz NF, Manchester M, Stuhlmann H, et al. Hydrazone ligation strategy to assemble multifunctional viral nanoparticles for cell imaging and tumor targeting. Nano Lett. 2010; 10:1093–7.
7). Hayden CM, Mackenzie AM, Skotnicki ML, Gibbs A. Turnip yellow mosaic virus isolates with experimentally produced recombinant virion proteins. J Gen Virol. 1998; 79:395–403.

8). Powel JD, Barbar E, Dreher TW. Turnip yellow mosaic virus forms infectious particles without the native beta-annulus structure and flexible coat protein N-terminus. Virology. 2012; 422:165–73.

9). Shin H-I, Chae K-H, Cho T-J. Modification of Turnip yellow mosaic virus coat protein and its effect on virion assembly. BMB reports. 2013; 46:495–500.

10). Verdurmen WPR, Brock R. Biological responses towards cationic peptides and drug carriers. Trends Pharmacol Sci. 2011; 32:116–24.

11). Cho T-J, Dreher TW. Encapsidation of genomic but not subgenomic Turnip yellow mosaic virus RNA by coat protein provided in trans. Virology. 2006; 356:126–35.

13). Shin H-I, Cho T-J. Read-through mutation in the coat protein ORF suppresses Turnip yellow mosaic virus subgenomic RNA accumulation. J Bacteriol Virol. 2013; 43:54–63.

14). Shin H-I, Tzanetakis IE, Dreher TW, Cho T-J. The 5′-UTR of Turnip yellow mosaic virus does not include a critical encapsidation signal. Virology. 2009; 387:427–35.

15). Deiman BALM, Verlaan PWG, Pleij CWA. In vitro transcription by the turnip yellow mosaic virus RNA polymerase: a comparison with the alfalfa mosaic virus and brome mosaic virus replicases. J Virol. 2000; 74:264–71.
16). Singh RN, Dreher TW. Specific site selection in RNA resulting from a combination of nonspecific secondary structure and -CCRboxes: initiation of minus strand synthesis by turnip yellow mosaic virus RNA-dependent RNA polymerase. RNA. 1998; 4:1083–95.
17). Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature. 2014; 511:366–70.

18). Rao ALN. Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol. 2006; 44:61–87.

Go to : 
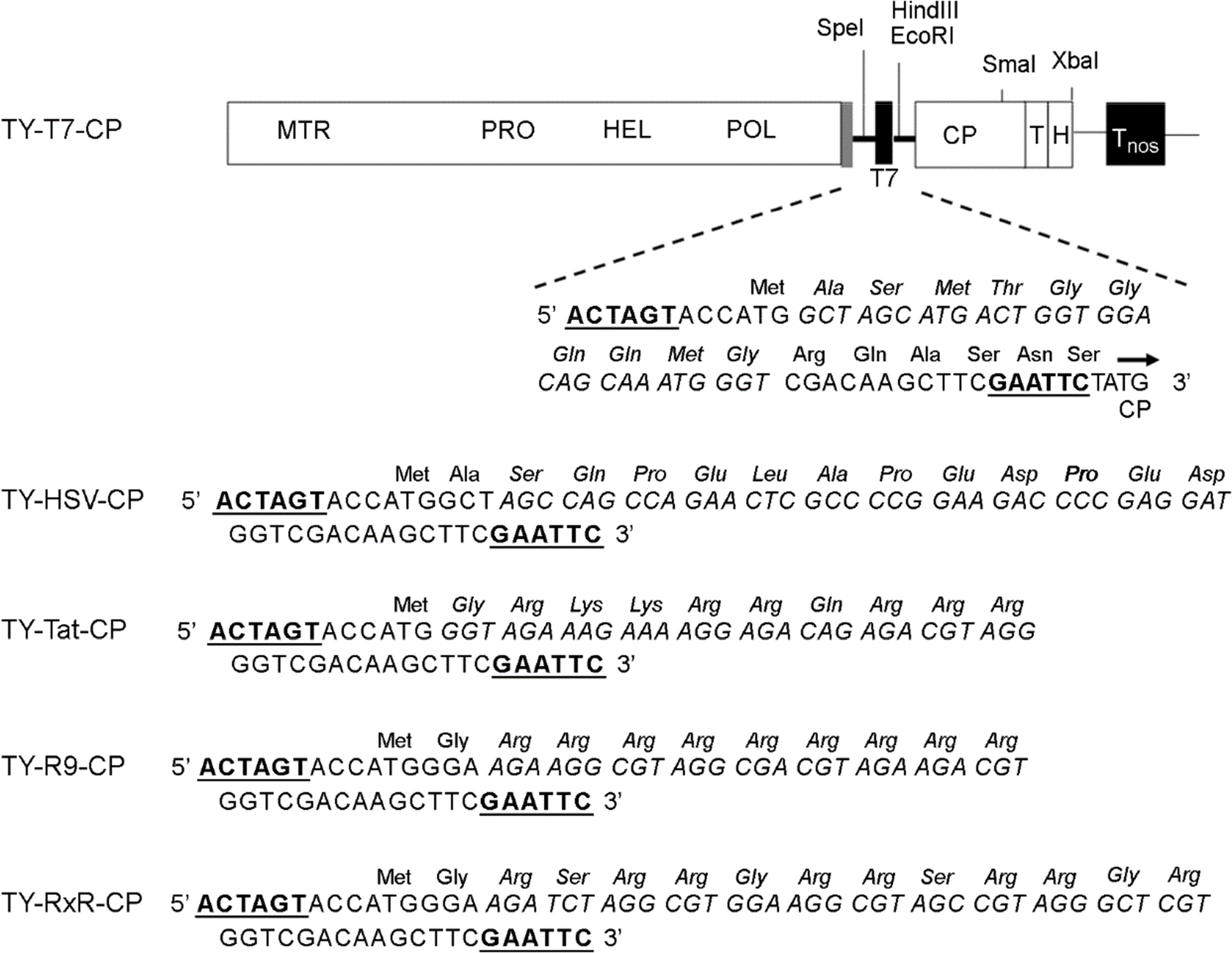 | Figure 1.TYMV constructs. TYMV CP was modified by inserting an oligonucleotide linker corresponding to the T7, HSV, Tat, (Arg)9, or (RxR)4 peptide into the Spe I/ Eco RI sites (underlined) of TY-M-CP (9). The nucleotide and amino acid sequences of the peptides are indicated in italic. The methyltransferase (MTR), protease (PRO) helicase (HEL) and polymerase (POL) domains of TYMV genome are shown. T and H represent the tRNA-like structure and HDV (hepatitis delta virus) ribozyme, respectively. Tnos is the transcription terminator of the nos gene. The sgRNA promoter, tymobox, is represented by grey box. |
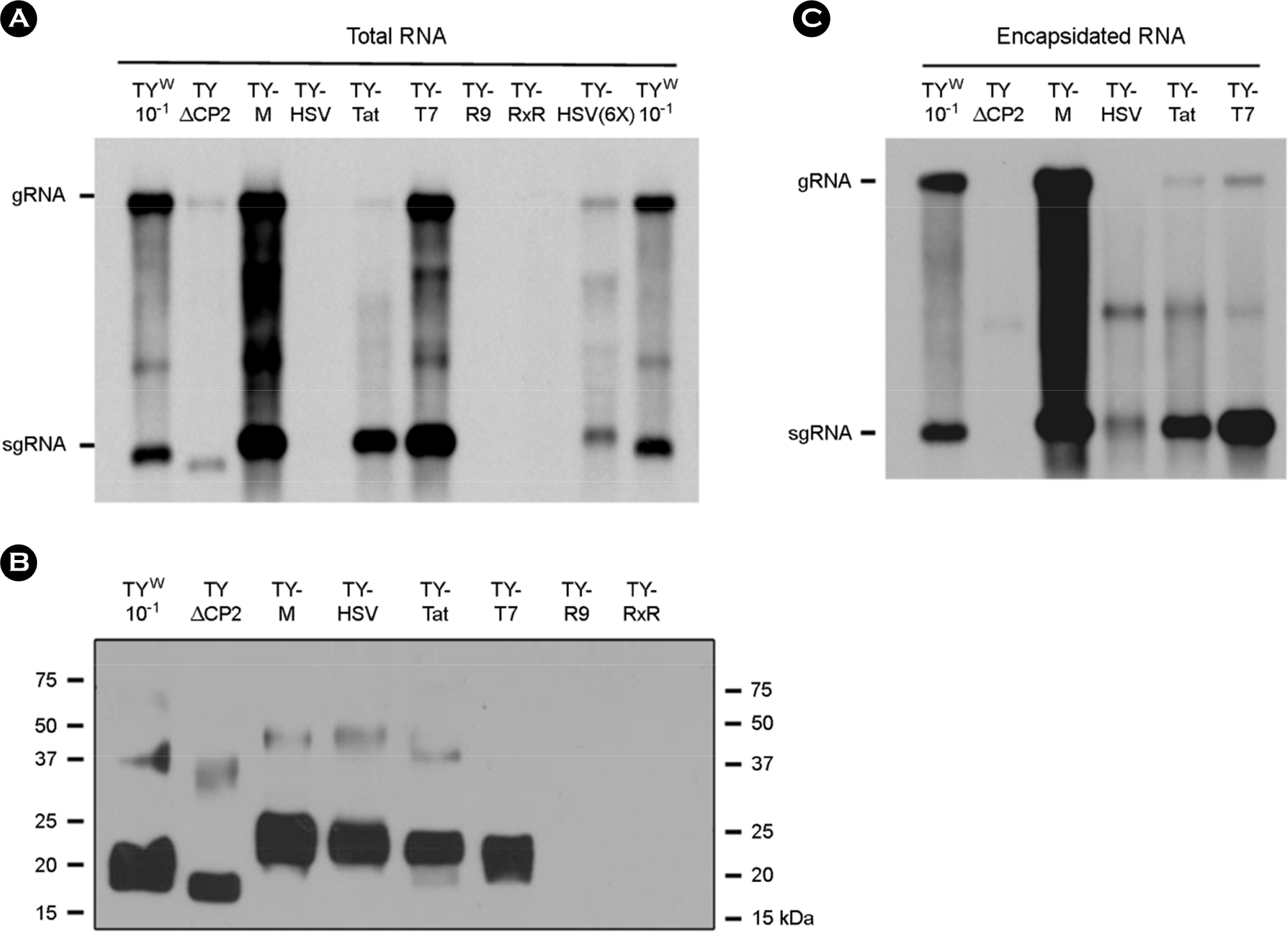 | Figure 2.Effect of the CP modification on replication and RNA packaging. (A and C) Northern analysis of total (A) and encapsidated (C) RNA. RNA samples were prepared from the N. benthamiana leaves collected 7 days after agroinfiltration. Viral RNAs were analyzed by Northern blot hybridization using the DIG-labeled probe representing the CP ORF. The positions of gRNA and sgRNA are indicated. (B) CP expression analysis. Leaf extracts were examined by SDS-PAGE, then Western blotting using anti-TYMV CP antibody and HRP-conjugated secondary antibody. TY W represents a wild-type TYMV construct. TYΔCP2, TY-T7, TY-HSV, TY-Tat, TY-R9, and TY-RxR represent the TYΔCP2+SL2, TY-T7-CP, TY-HSV-CP, TY-Tat-CP, TY-R9-CP, and TY-RxR-CP constructs, respectively. |
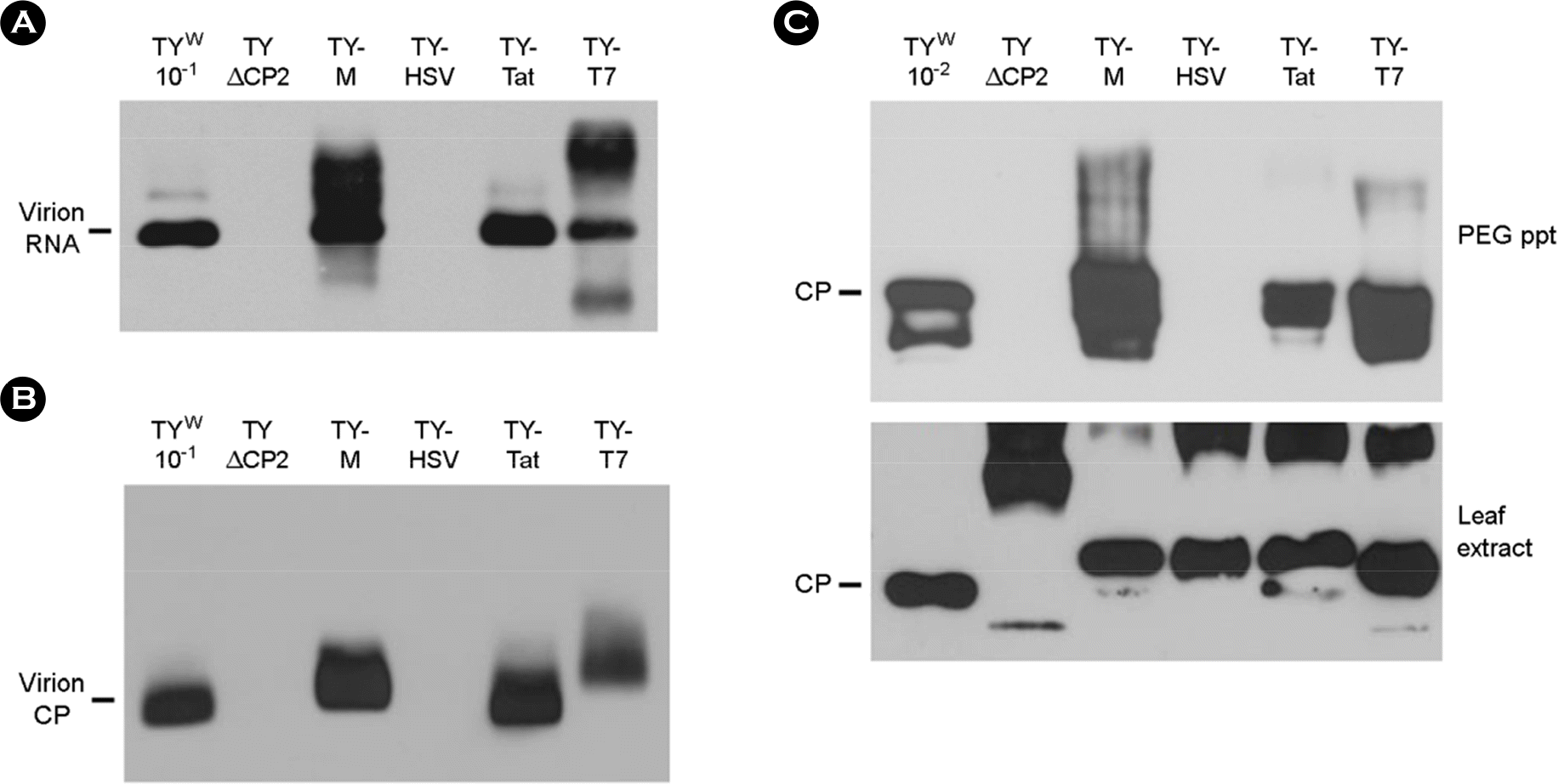 | Figure 3.Mobility shift assay of virions. (A) Examination of virion RNA by Northern analysis. PEG-precipitated virions were electrophoresed on a 1% agarose gel and then treated with 0.2 N NaOH. The liberated virion RNA was blotted onto nylon membrane and examined by hybridization with a DIG-labeled CP DNA probe. (B) Examination of virions by Western analysis. After non-denaturing agarose gel electrophoresis, the virions were transferred to nitrocellulose membrane, and were probed with anti-TYMV CP. (C) Western analysis of CP after SDS-PAGE. CPs in PEG-precipitated samples (top panel) and in leaf extracts (bottom panel) were examined as described in Figure 2C. |
 | Figure 4.Assembly of the modified CPs expressed from CP constructs. (A) A CP construct expressing recombinant CPs. T7-CP is shown as a representative CP construct. Ω represents a translational enhancer from Tobacco mosaic virus. Virions in leaf extracts (B) or virions precipitated with PEG (C) were electrophoresed on a 1% agarose gel, and were examined by Western blot analysis (top panels), as described in Figure 3B. CPs in the samples were also examined by Western analysis after SDS-PAGE and blotting to nitrocellulose membrane (bottom panels). CP was detected as described in Figure 2C. CPpA, described previously (9), produces unmodified TYMV CPs. |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download