Abstract
HilA is a central regulator of Salmonella pathogenicity island 1 (SPI1), which is necessary for host invasion by Salmonella and induction of gastroenteritis. The iagB lies downstream of hilA and is thought to be co-transcribed with hilA, but iagB expression has not yet been analyzed directly. In this study, iagB expression in various mutant strains was measured to determine whether the expression pattern was similar to that of hilA. A β-galactosidase assay revealed that iagB expression was greater under shaking than standing culture condition. iagB expression was decreased in relA/ spoT and ihfB mutants but not in luxS mutant, in line with previous reports on hilA expression. The hilA and iagB mRNA levels decreased by approximately 2-fold in arcA mutant grown aerobically and increased by approximately 10-fold in fnr mutant grown anaerobically. Although the fold changes in hilA and iagB mRNA level differed in hfq mutant strain, the patterns of timeand Hfq-dependent regulation were similar for both genes. Thus, iagB and hilA exhibited similar expression patterns in various mutational backgrounds and under different growth condition.
Go to : 
REFERENCES
1). Cotter PA, DiRita VJ. Bacterial virulence gene regulation: an evolutionary perspective. Annu Rev Mcrobiol. 2000; 54:519–65.

2). Morschhäuser J, Köhler G, Ziebuhr W, Blum-Oehler G, Dobrindt U, Hacker J. Evolution of microbial pathogens. Philos Trans R Soc Lond B Biol Sci. 2000; 355:695–704.

3). Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001; 52:259–74.
4). Wain J, House D, Pickard D, Dougan G, Frankel G. Acquisition of virulence-associated factors by the enteric pathogens Escherichia coli and Salmonella enterica. Philos Trans R Soc Lond B Biol Sci. 2001; 356:1027–34.
5). Jones BD. Salmonella invasion gene regulation: A story of environmental awareness. J Microbiol. 2005; 43:110–7.
6). Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Micorbiol. 2007; 10:24–9.
7). Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics. 2012; 190:79–90.
8). Baxter MA, Fahlen TF, Wilson RL, Jones BD. HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect Immun. 2003; 71:1295–305.
9). Bajaj V, Hwang C, Lee CA. hilA is a novel OmpR/ToxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995; 18:715–27.
10). Boddicker JD, Knosp BM, Jones BD. Transcription of the Salmonella invasion gene activator, hilA, requires HilD activation in the absence of negative regulators. J Bacteriol. 2003; 185:525–33.
11). Ellermeier CD, Ellermeier JR, Slauch JM. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2005; 57:691–705.
12). Eichelberg K, Hardt WD, Galán JE. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol Microbiol. 1999; 33:139–52.
13). Rakeman JL, Bonifield HR, Miller SI. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J Bacteriol. 1999; 181:3096.
14). Schechter LM, Damrauer SM, Lee CA. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999; 32:629–42.
15). Olekhnovich IN, Kadner RJ. DAN-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol. 2002; 184:4148–60.
16). Lucas RL, Lee CA. Roles of hilC and hilD in regulation of hilA expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2001; 183:2733–45.
17). Lim S, Yong K, Ryu S. Analysis of Salmonella pathogenicity island 1 expression in response to the changes of osmolarity. J Microbiol Biotechnol. 2005; 15:175–82.
18). De Keersmaecker SC, Marchal K, Verhoven TL, Engelen K, Vanderleyden J, Detweiler CS. Microarray analysis and motif detection reveal new targets of the Salmonella enterica serovar Typhimurium HilA regulatory protein, including hilA itself. J Bacteriol. 2005; 187:4381–91.
19). Lucas RL, Lostroh CP, DiRusso CC, Spector MP, Wanner BL, Lee CA. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000; 182:1872–82.
20). Sukhan A, Kubori T, Wilson J, Galán JE. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol. 2001; 183:1159–67.
21). Felise HB, Nguyen HV, Pfuetzner RA, Barry KC, Jackson SR, Blanc MP, et al. An inhibitor of Gram-negative bacterial virulence protein secretion. Cell Host Microbe. 2008; 4:325–36.

22). Lim S, Yoon H, Kim M, Han A, Choi J, Choi J, et al. Hfq and ArcA are involved in the stationary phase-dependent activation of Salmonella pathogenicity island 1 (SPI1) under shaking culture conditions. J Microbiol Biotechnol. 2013; 23:1664–72.
23). Maloy SR, Stewart VJ, Taylor RK. Genetic analysis of pathogenic bacteria: A laboratory manual. Plainview, N.Y.: Cold Spring Harbor Laboratory Press;1996.
24). Lim S, Lee B, Kim M, Kim D, Yoon H, Yong K, et al. Analysis of HilC/D-dependent invF promoter expression under different culture conditions. Microb Pathog. 2012; 52:359–66.
25). Choi J, Shin D, Ryu S. Implication of quorum sensing in Salmonella enterica serovar Typhimurium virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect Immun. 2007; 75:4885–90.
26). Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, et al. ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem. 2004; 279:34183–90.
27). Queiroz MH, Madrid C, Paytubi S, Balsalobre C, Juárez A. Integration host factor alleviates H-NS silencing of the Salmonella enterica serovar Typhimurium master regulator of SPI1, hilA. Microbiology. 2011; 157:2504–14.
28). Evans MR, Fink RC, Vazquez-Torres A, Porwollik S, Jones-Carson J, McClelland M, et al. Analysis of the ArcA regulon in anaerobically grown Salmonella enterica sv. Typhimurium. BMC Microbiol. 2011; 11:58.

29). Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, et al. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J Bacteriol. 2007; 189:2262–73.
30). Van Immerseel F, Eeckhaut V, Boyen F, Pasmans F, Haesebrouck F, Ducatelle R. Mutations influencing expression of the Salmonella enterica serovar Enteritidis pathogenicity island I key regulator hilA. Antonie Van Leeuwenhoek. 2008; 94:455–61.
31). Lim S, Choi J, Kim M, Yoon H. Temporal regulation of Salmonella pathogenicity Island 1 (SPI-1) hilA by Hfq in Salmonella enterica serovar typhimurium. J Korean Soc Appl Biol Chem. 2015; 58:169–72.
32). Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, et al. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008; 4:e1000163.

33). Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol. 2007; 63:193–217.
34). Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, et al. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 2016; 35:991–1011.
Go to : 
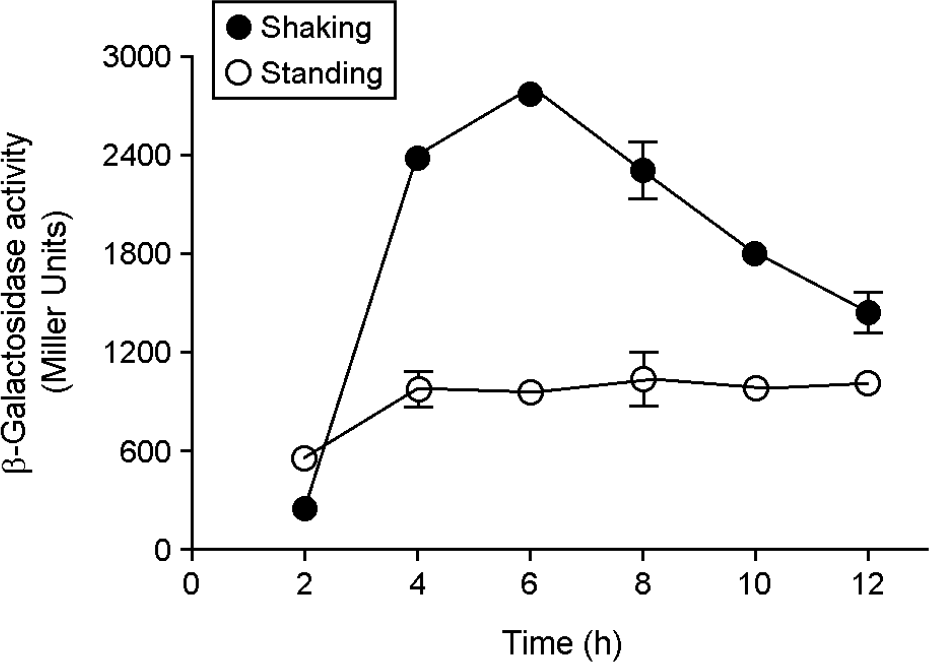 | Figure 1.
Transcriptional analysis of iagB under different growth conditions. The S. Typhimurium strain harboring a chromosomal iagB:: lacZY fusion was grown in LB medium under shaking (●) or standing conditions (○). The expression level was determined by measuring the β-galactosidase activity (Miller units) at the indicated times. Data are presented as the mean ± standard error of three independent experiments performed in duplicate. |
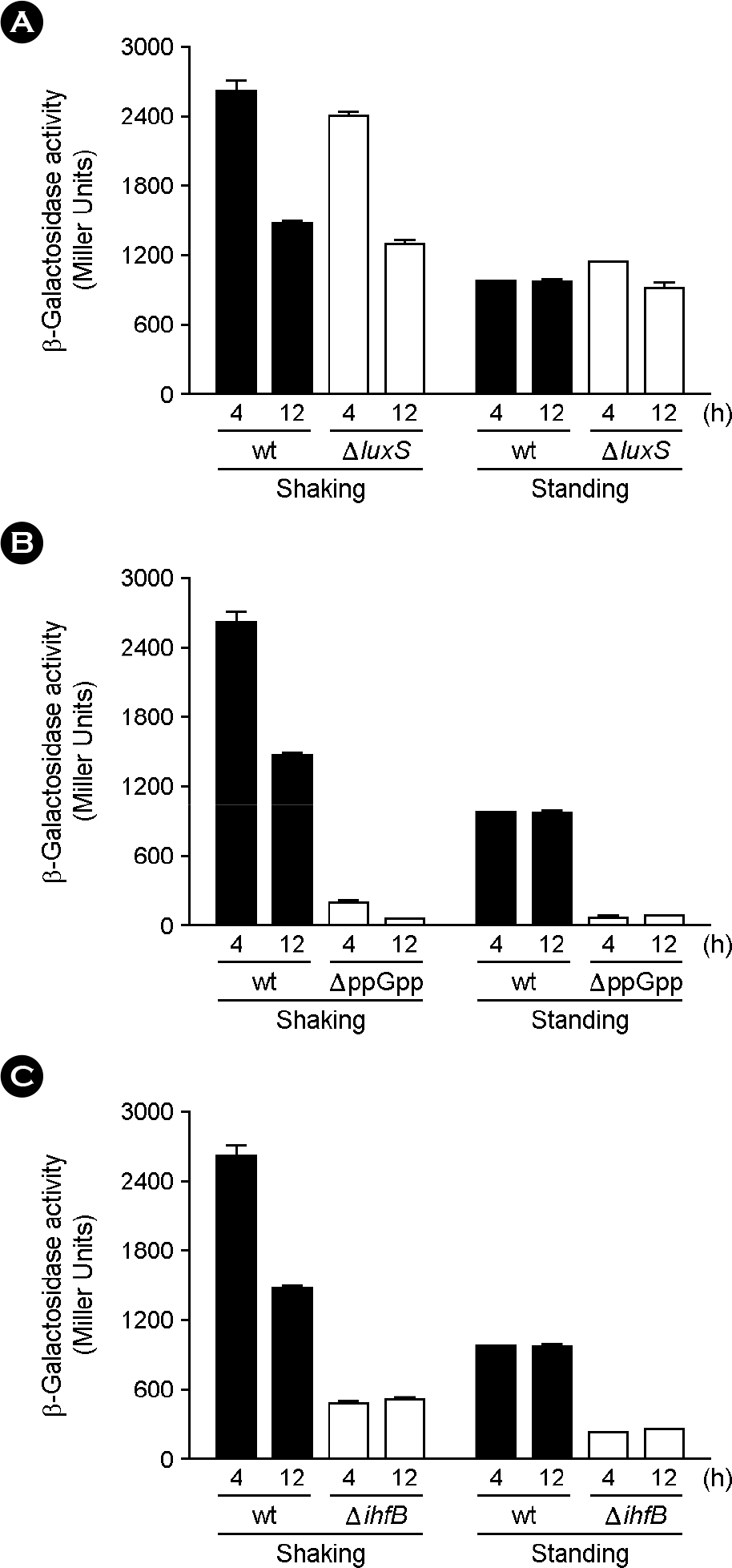 | Figure 2.
Transcriptional analysis of iagB in different mutant strains. The β-galactosidase activities (Miller units) of chromosomal iagB::lacZY were measured in ΔluxS (A), ΔppGpp (ΔrelA/ΔspoT)(B), and Δ ihfB (C) strains at 4 h p.i. (early stationary phase) and 12 h p.i. (late stationary phase) under shaking and standing culture conditions, respectively. Data are presented as the mean ± standard error of three independent experiments performed in duplicate. |
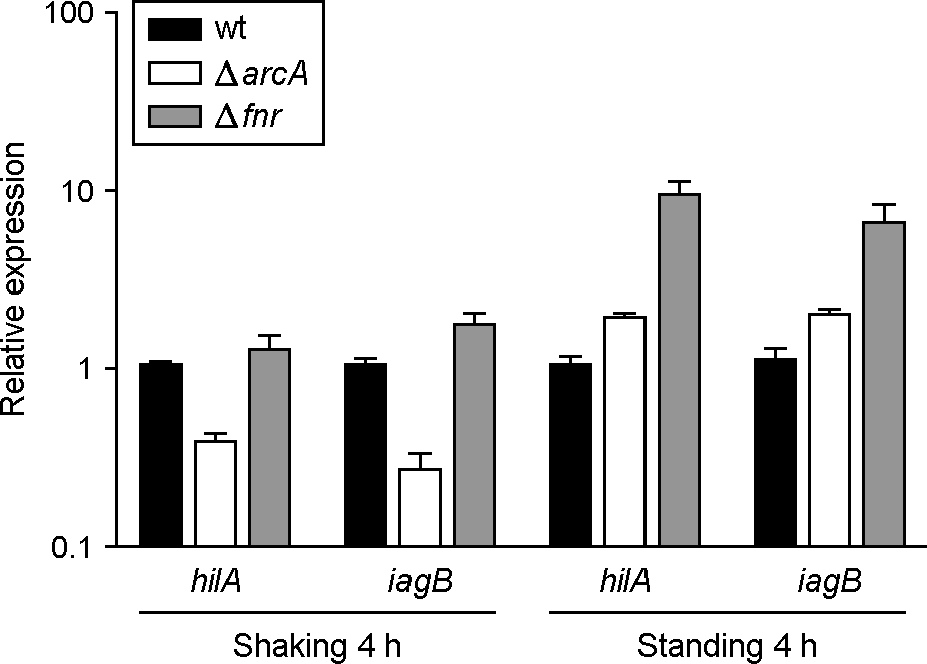 | Figure 3.
Transcriptional analysis of hilA and iagB in arcA and fnr mutants. The mRNA levels of hilA and iagB were determined by qRT-PCR. Total RNA was isolated from strains grown for 4 h under shaking and standing culture conditions. The values for the relative expression were determined by defining the mRNA levels from the wild-type strain as 1. The expression levels of the target genes were normalized to 16S rRNA gene. Data are presented as the mean ± standard error of three independent experiments performed in duplicate. |
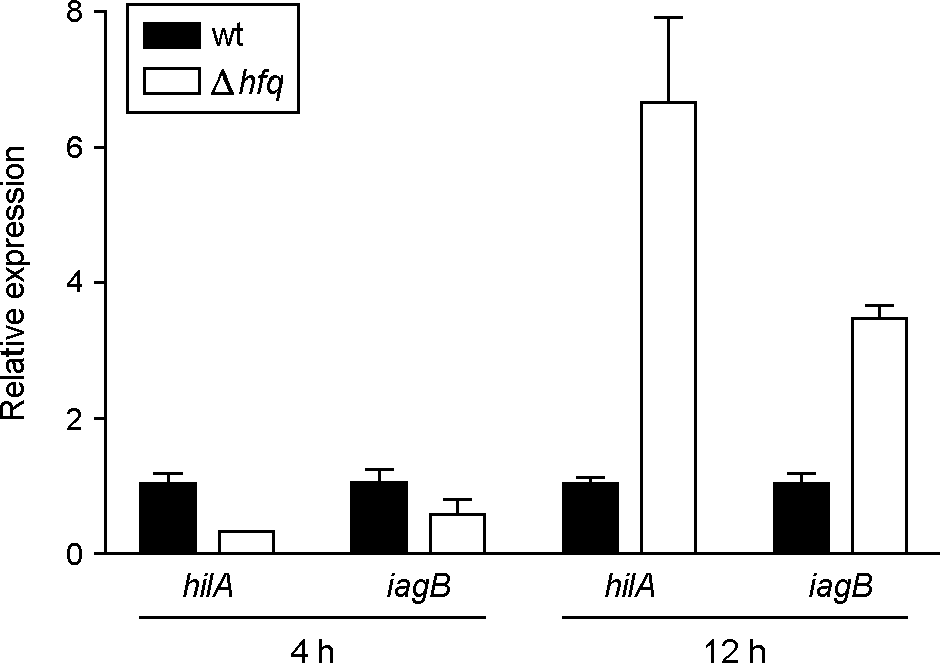 | Figure 4.
Transcriptional analysis of hilA and iagB in hfq mutants. The mRNA levels of hilA and iagB were determined by qRT-PCR. Total RNA was isolated from strains grown for 4 h and 12 h under shaking culture conditions. The values for the relative expression were determined by defining the mRNA levels from the wild-type strain as 1. The expression levels of the target genes were normalized to 16S rRNA gene. Data are presented as the mean ± standard error of three independent experiments performed in duplicate. |
Table 1.
The bacterial strains




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download