Abstract
Since 1990 when Wolff and co-authors proved that both RNA and DNA expression vectors containing interest gene were directly injected into mouse muscle and expressed the protein in vivo, the concept of gene vaccine has been broadly tested in the vaccine field. However, due to the limitations of technology and the misconception about RNA, most previous studies have focused on the DNA vaccine. Recently, the RNA vaccine has emerged as a new game-changing disruptive innovation technology in the vaccine field. This review has covered the characteristics of the RNA vaccine, including its strengths and weaknesses. Finally, we have suggested future directions for the RNA vaccine.
Go to : 
REFERENCES
1). Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990; 247:1465–8.
3). Ferraro B, Morrow MP, Hutnick NA, Shin TH, Lucke CE, Weiner DB. Clinical applications of DNA vaccines: current progress. Clin Infect Dis. 2011; 53:296–302.

4). Liu MA. DNA vaccines: an historical perspective and view to the future. Immunol Rev. 2011; 239:62–84.

6). Hilleman MR. Recombinant vector vaccines in vaccinology. Dev Biol Stand. 1994; 82:3–20.
7). Sayour EJ, Sanchez-Perez L, Flores C, Mitchell DA. Bridging infectious disease vaccines with cancer immunotherapy: a role for targeted RNA based immunotherapeutics. J Immunother Cancer. 2015; 3:13.

8). Leitner WW. Immune-Activating Mechanisms of Replicase-Based DNA and RNA Vaccines and Their Role in Immune-Apoptosis. Springer;Vienna. 2012. p. 67–88.
9). Geall AJ, Mandl CW, Ulmer JB. RNA: the new revolution in nucleic acid vaccines. Semin Immunol. 2013; 25:152–9.

10). Halstead SB, Thomas SJ. New Japanese encephalitis vaccines: alternatives to production in mouse brain. Expert Rev Vaccines. 2011; 10:355–64.

11). Widera G, Austin M, Rabussay D, Goldbeck C, Barnett SW, Chen M, et al. Increased DNA vaccine delivery and immunogenicity by electroporation in vivo. J Immunol. 2000; 164:4635–40.
12). Petsch B, Schnee M, Vogel AB, Lange E, Hoffmann B, Voss D, et al. Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol. 2012; 30:1210–6.
13). Probst J, Weide B, Scheel B, Pichler BJ, Hoerr I, Rammensee HG, et al. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007; 14:1175–80.
14). Roesler E, Weiss R, Weinberger EE, Fruehwirth A, Stoecklinger A, Mostböck S, et al. Immunize and disappear-safety-optimized mRNA vaccination with a panel of 29 allergens. J Allergy Clin Immunol. 2009; 124:1070–7.

15). Dormitzer PR, Ulmer JB, Rappuoli R. Structure-based antigen design: a strategy for next generation vaccines. Trends Biotechnol. 2008; 26:659–67.

16). Ulmer JB, Valley U, Rappuoli R. Vaccine manufacturing: challenges and solutions. Nat Biotechnol. 2006; 24:1377–83.

19). Pasquinelli AE, Dahlberg JE, Lund E. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA. 1995; 1:957–67.
20). Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE. Synthesis and properties of mRNAs containing the novel "anti-reverse" cap analogs 7-methyl(3′-O-methyl)- GpppG and 7-methyl (3′-deoxy)GpppG. RNA. 2001; 7:1486–95.
21). Venkatesan S, Gershowitz A, Moss B. Modification of the 5′ end of mRNA. Association of RNA triphosphatase with the RNA guanylyltransferase-RNA (guanine-7-)methyl- transferase complex from vaccinia virus. J Biol Chem. 1980; 255:903–8.
22). Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE. Differential inhibition of mRNA degradation pathways by novel cap analogs. J Biol Chem. 2006; 281:1857–67.

23). Zohra FT, Chowdhury EH, Tada S, Hoshiba T, Akaike T. Effective delivery with enhanced translational activity synergistically accelerates mRNA-based transfection. Biochem Biophys Res Commun. 2007; 358:373–8.

24). Rydzik AM, Kulis M, Lukaszewicz M, Kowalska J, Zuberek J, Darzynkiewicz ZM, et al. Synthesis and properties of mRNA cap analogs containing imidodiphosphate moiety– fairly mimicking natural cap structure, yet resistant to enzymatic hydrolysis. Bioorg Med Chem. 2012; 20:1699–710.
25). Grudzien-Nogalska E, Jemielity J, Kowalska J, Darzynkiewicz E, Rhoads RE. Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA. 2007; 13:1745–55.

26). IRESite. The database of experimentally verified IRES structures. http://iresite.org/.
27). Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem. 2010; 285:29033–8.

28). Li H, Jiang JD, Peng ZG. MicroRNA-mediated interactions between host and hepatitis C virus. World J Gastroenterol. 2016; 22:1487–96.

29). Scheel TK, Luna JM, Liniger M, Nishiuchi E, Rozen-Gagnon K, Shlomai A, et al. A Broad RNA Virus Survey Reveals Both miRNA Dependence and Functional Sequestration. Cell Host Microbe. 2016; 19:409–23.

30). Mishima Y, Tomari Y. Codon Usage and 3′ UTR Length Determine Maternal mRNA Stability in Zebrafish. Mol Cell. 2016; 61:874–85.

31). Peng J, Schoenberg DR. mRNA with a <20-nt poly(A) tail imparted by the poly(A)-limiting element is translated as efficiently in vivo as long poly(A) mRNA. RNA. 2005; 11:1131–40.
32). Zhao Y, Moon E, Carpenito C, Paulos CM, Liu X, Brennan AL, et al. Multiple injections of electroporated autologous T cells expressing a chimeric antigen receptor mediate regression of human disseminated tumor. Cancer Res. 2010; 70:9053–61.

33). Kreiter S, Selmi A, Diken M, Sebastian M, Osterloh P, Schild H, et al. Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J Immunol. 2008; 180:309–18.

34). Schlake T, Thess A, Fotin-Mleczek M, Kallen KJ. Developing mRNA-vaccine technologies. RNA Biol. 2012; 9:1319–30.

35). Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 2001; 53:290–8.

36). Duret L. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 2002; 12:640–9.

37). Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986; 44:283–92.

38). Liu Q. Comparative analysis of base biases around the stop codons in six eukaryotes. Biosystems. 2005; 81:281–9.

39). Lundstrom K. Self-replicating RNA viral vectors in vaccine development and gene therapy. Future Virol. 2016; 11:345–56.

40). Kim HJ, Kim A, Miyata K, Kataoka K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv Drug Deliv Rev. 2016; 104:61–77.

41). Leitner WW, Ying H, Driver DA, Dubensky TW, Restifo NP. Enhancement of tumor-specific immune response with plasmid DNA replicon vectors. Cancer Res. 2000; 60:51–5.
42). Scheel B, Braedel S, Probst J, Carralot JP, Wagner H, Schild H, et al. Immunostimulating capacities of stabilized RNA molecules. Eur J Immunol. 2004; 34:537–47.

43). Scheel B, Teufel R, Probst J, Carralot JP, Geginat J, Radsak M, et al. Toll-like receptor-dependent activation of several human blood cell types by protamine-condensed mRNA. Eur J Immunol. 2005; 35:1557–66.

44). Rettig L, Haen SP, Bittermann AG, von Boehmer L, Curioni A, Krämer SD, et al. Particle size and activation threshold: a new dimension of danger signaling. Blood. 2010; 115:4533–41.

45). Isaacs A, Cox RA, Rotem Z. Foreign nucleic acids as the stimulus to make interferon. Lancet. 1963; 2:113–6.

46). Field AK, Tytell AA, Lampson GP, Hilleman MR. Inducers of interferon and host resistance. II. Multistranded synthetic polynucleotide complexes. Proc Natl Acad Sci U S A. 1967; 58:1004–10.

47). Wilkins C, Gale M Jr. Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010; 22:41–7.

48). Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007; 318:1455–8.

49). Hovanessian AG. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: the 2′-5′oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev. 2007; 18:351–61.

50). Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006; 441:101–5.

51). Liljeström P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology (N Y). 1991; 9:1356–61.

52). Martinon F, Krishnan S, Lenzen G, Magné R, Gomard E, Guillet JG, et al. Induction of virus-specific cytotoxic T lymphocytes in vivo by liposome-entrapped mRNA. Eur J Immunol. 1993; 23:1719–22.
53). Krieg PA, Melton DA. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984; 12:7057–70.
54). Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984; 12:7035–56.
55). Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987; 15:8783–98.

56). Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci U S A. 2012; 109:14604–9.

57). Piggott JM, Sheahan BJ, Soden DM, O'Sullivan GC, Atkins GJ. Electroporation of RNA stimulates immunity to an encoded reporter gene in mice. Mol Med Rep. 2009; 2:753–6.

58). Carralot JP, Probst J, Hoerr I, Scheel B, Teufel R, Jung G, et al. Polarization of immunity induced by direct injection of naked sequence-stabilized mRNA vaccines. Cell Mol Life Sci. 2004; 61:2418–24.

59). Granstein RD, Ding W, Ozawa H. Induction of antitumor immunity with epidermal cells pulsed with tumor-derived RNA or intradermal administration of RNA. J Invest Dermatol. 2000; 114:632–6.

60). Johansson DX, Ljungberg K, Kakoulidou M, Liljeström P. Intradermal electroporation of naked replicon RNA elicits strong immune responses. PLoS One. 2012; 7:e29732.

61). Zhou WZ, Hoon DS, Huang SK, Fujii S, Hashimoto K, Morishita R, et al. RNA melanoma vaccine: induction of antitumor immunity by human glycoprotein 100 mRNA immunization. Hum Gene Ther. 1999; 10:2719–24.

62). Kreiter S, Selmi A, Diken M, Koslowski M, Britten CM, Huber C, et al. Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res. 2010; 70:9031–40.

63). Kreiter S, Diken M, Selmi A, Türeci Ö, Sahin U. Tumor vaccination using messenger RNA: prospects of a future therapy. Curr Opin Immunol. 2011; 23:399–406.

64). Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Current prospects for mRNA gene delivery. Eur J Pharm Biopharm. 2009; 71:484–9.

65). Qiu P, Ziegelhoffer P, Sun J, Yang NS. Gene gun delivery of mRNA in situ results in efficient transgene expression and genetic immunization. Gene Ther. 1996; 3:262–8.
66). Steitz J, Britten CM, Wölfel T, Tüting T. Effective induction of anti-melanoma immunity following genetic vaccination with synthetic mRNA coding for the fusion protein EGFP.TRP2. Cancer Immunol Immunother. 2006; 55:246–53.

67). Evans RK, Xu Z, Bohannon KE, Wang B, Bruner MW, Volkin DB. Evaluation of degradation pathways for plasmid DNA in pharmaceutical formulations via accelerated stability studies. J Pharm Sci. 2000; 89:76–87.

Go to : 
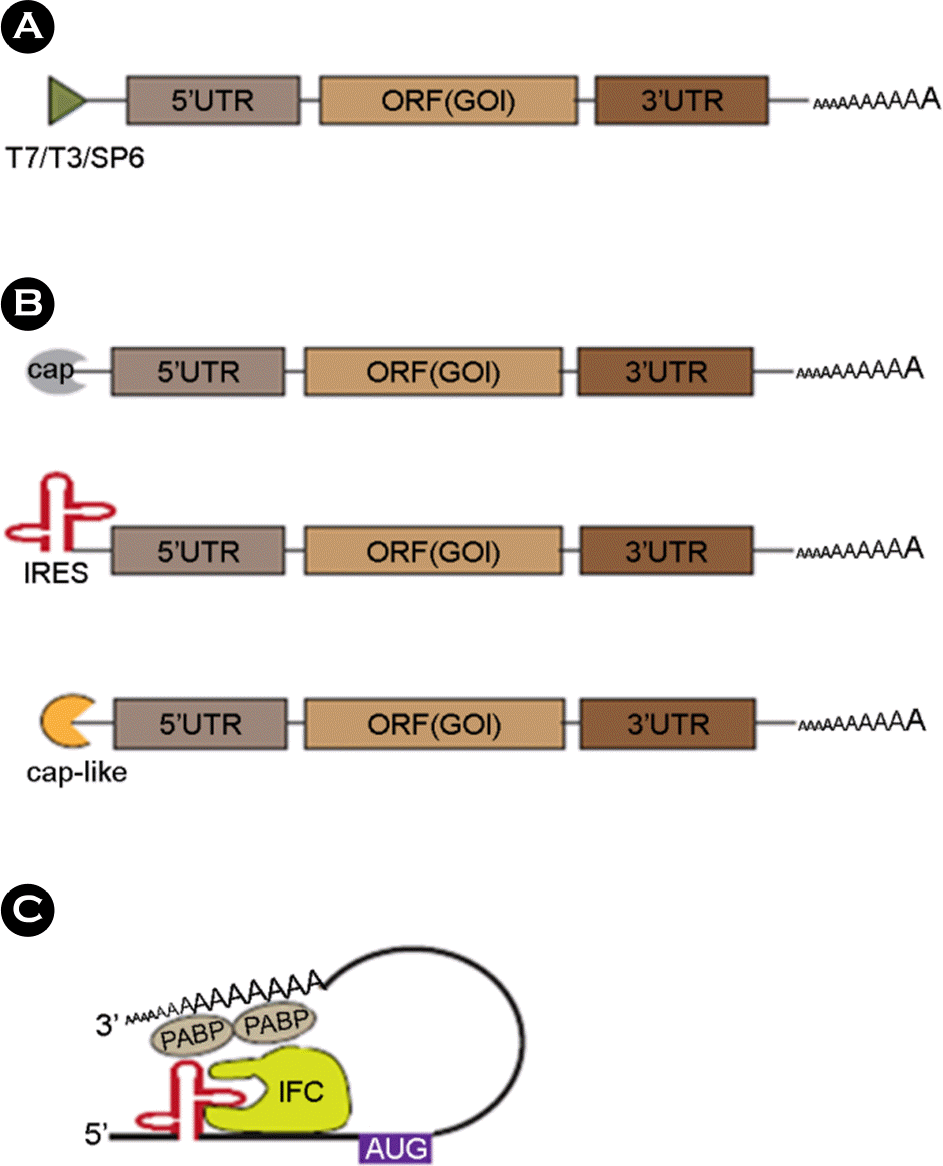 | Figure 1.Different mechanism between DNA vaccines and the RNA vaccines makes different features such as efficiency and stability. |
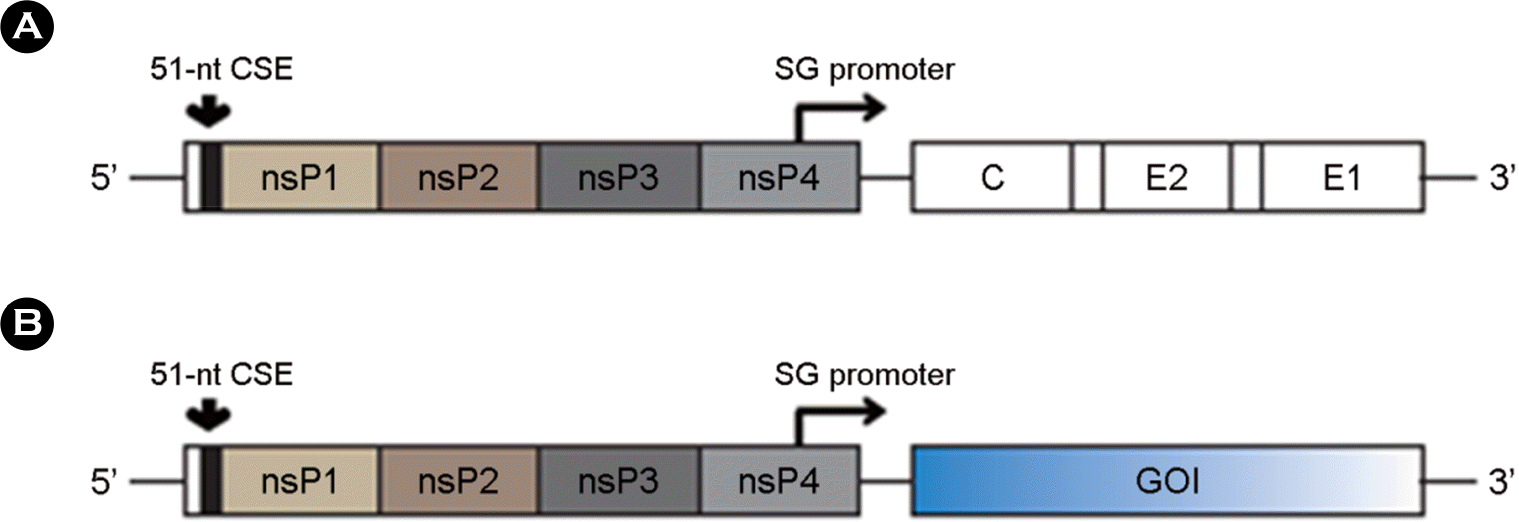 | Figure 2.(A) RNA vaccine contain in 5′-UTR region, ORF (Open Reading Frame, Gene of Interest, GOI) and 3′-UTR region. (B) Various modifications on 5′-UTR region of vaccines are essential for initiation of translation. IRES: Internal Ribosome Entry site, Cap-like: Cap-like structures. (C) Translation of some mRNAs depends on interactions between 5′-UTR and 3′-UTR region. PABP: Poly A Binding Protein, IFC: a Complex of translation Initiation Factors. |
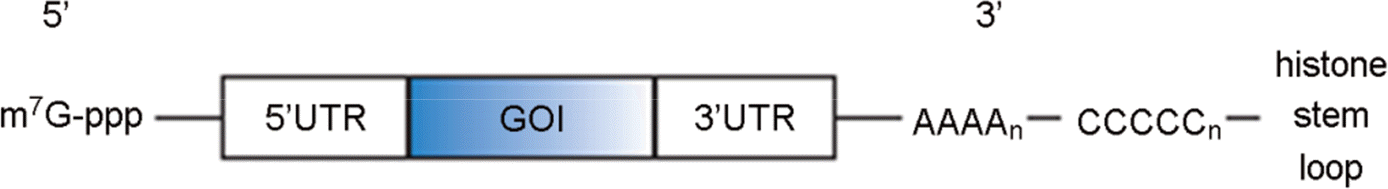 | Figure 3.(A) Schematic structure of self-replicon RNA vaccine contains in structural and nonstructural protein coding region to replicate.51-nt CSE: 51-nt Conserved Sequence Element, SG promoter: sub-genomic promoter (B) Replacement of structural protein coding region with GOI (Gene of Interest). |




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download