Abstract
Nuclear targeting of bacterial proteins in host cells and subsequent interaction with nuclear molecules are an emerging pathogenic mechanism of bacteria. In this study, we predicted the nuclear targeting proteins with nuclear localization signals (NLSs) in Staphylococcus aureus using bioinformatic analysis. A total of 51 proteins of S. aureus, comprising of 24 functional and 27 hypothetical proteins, were predicted to carry putative NLSs. Among them, β-lactamase and MsrR proteins with the putative NLSs were selected to determine the nuclear targeting in host cells. Fusion proteins of BlaZ-green fluorescent protein (GFP) were evenly distributed in the nuclei of host cells and subsequently induced host cell death. However, fusion proteins of MsrR-GFP were not localized in the nuclei of host cells In conclusion, screening of nuclear targeting proteins with NLSs and determination of their pathology in host cells may open up the new field of S. aureus pathogenesis.
Go to : 
REFERENCES
1). Matarrese P, Falzano L, Fabbri A, Gambardella L, Frank C, Geny B, et al. Clostridium difficile toxin B causes apoptosis in epithelial cells by thrilling mitochondria. Involvement of ATP-sensitive mitochondrial potassium channels. J Biol Chem. 2007; 282:9029–41.
2). Müller A, Günther D, Brinkmann V, Hurwitz R, Meyer TF, Rudel T. Targeting of the pro-apoptotic VDAC-like porin (PorB) of Neisseria gonorrhoeae to mitochondria of infected cells. EMBO J. 2000; 19:5332–43.
3). Nagai T, Abe A, Sasakawa C. Targeting of enteropathogenic Escherichia coli EspF to host mitochondria is essential for bacterial pathogenesis: critical role of the 16th leucine residue in EspF. J Biol Chem. 2005; 280:2998–3011.
4). Nougayrède JP, Donnenberg MS. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell Microbiol. 2004; 6:1097–111.
5). Papatheodorou P, Domańska G, Oxle M, Mathieu J, Selchow O, Kenny B, et al. The enteropathogenic Escherichia coli (EPEC) Map effector is imported into the mitochondrial matrix by the TOM/Hsp70 system and alters organelle morphology. Cell Microbiol. 2006; 8:677–89.
7). Elwell C, Chao K, Patel K, Dreyfus L. Escherichia coli CdtB mediates cytolethal distending toxin cell cycle arrest. Infect Immun. 2001; 69:3418–22.
8). Toyotome T, Suzuki T, Kuwae A, Nonaka T, Fukuda H, Imajoh-Ohmi S, et al. Shigella protein IpaH (9.8) is secreted from bacteria within mammalian cells and transported to the nucleus. J Biol Chem. 2001; 276:32071–9.
9). Haraga A, Miller SI. A Salmonella enterica serovar Typhimurium translocated leucine-rich repeat effector protein inhibits NF-κB-dependent gene expression. Infect Immun. 2003; 71:4052–8.
10). Benabdillah R, Mota LJ, Lützelschwab S, Demoinet E, Cornelis GR. Identification of a nuclear targeting signal in YopM from Yersinia spp. Microb Pathog. 2004; 36:247–61.
11). Choi CH, Hyun SH, Lee JY, Lee JS, Lee YS, Kim SA, et al. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell Microbiol. 2008; 10:309–19.
12). Lee JC, Kim DS, Moon DC, Lee JH, Kim MJ, Lee SM, et al. Prediction of bacterial proteins carrying a nuclear localization signal and nuclear targeting of HsdM from Klebsiella pneumoniae. J Microbiol. 2009; 47:641–5.
13). Lara-Tejero M, Galán JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000; 290:354–7.

14). McSweeney LA, Dreyfus LA. Nuclear localization of the Escherichia coli cytolethal distending toxin CdtB subunit. Cell Microbiol. 2004; 6:447–58.
15). Nishikubo S, Ohara M, Ueno Y, Ikura M, Kurihara H, Komatsuzawa H, et al. An N-terminal segment of the active component of the bacterial genotoxin cytolethal distending toxin B (CDTB) directs CDTB into the nucleus. J Biol Chem. 2003; 278:50671–81.

16). Choi CH, Hyun SH, Kim J, Lee YC, Seol SY, Cho DT, et al. Nuclear translocation and DNAse I-like enzymatic activity of Acinetobacter baumannii outer membrane protein A. FEMS Microbiol Lett. 2008; 288:62–7.
17). Choi CH, Lee EY, Lee YC, Park TI, Kim HJ, Hyun SH, et al. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol. 2005; 7:1127–38.
18). Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998; 4:351–64.
19). Moroianu J. Distinct nuclear import and export pathways mediated by members of the karyopherin β family. J Cell Biochem. 1998; 70:231–9.

20). Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004; 14:547–56.

21). Pemberton LF, Blobel G, Rosenblum JS. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998; 10:392–9.

22). Moon DC, Gurung M, Lee JH, Lee YS, Choi CW, Kim SI, et al. Screening of nuclear targeting proteins in Acinetobacter baumannii based on nuclear localization signals. Res Microbiol. 2012; 163:279–85.
23). Lee JH, Jun SH, Baik SC, Kim DR, Park JY, Lee YS, et al. Prediction and screening of nuclear targeting proteins with nuclear localization signals in Helicobacter pylori. J Microbiol Methods. 2012; 91:490–6.
24). Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbour, New York: Cold Spring Harbour Press;1989.
25). Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, et al. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One. 2011; 6:e27958.
26). Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, et al. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics. 2009; 9:5425–36.
27). Lee J, Lee EY, Kim SH, Kim DK, Park KS, Kim KP, et al. Staphylococcus aureus extracellular vesicles carry biologically active β-lactamase. Antimicrob Agents Chemother. 2013; 57:2589–95.
28). Lee JC. Staphylococcus aureus membrane vesicles and its potential role in bacterial pathogenesis. J Bacteriol Virol. 2012; 42:181–8.
Go to : 
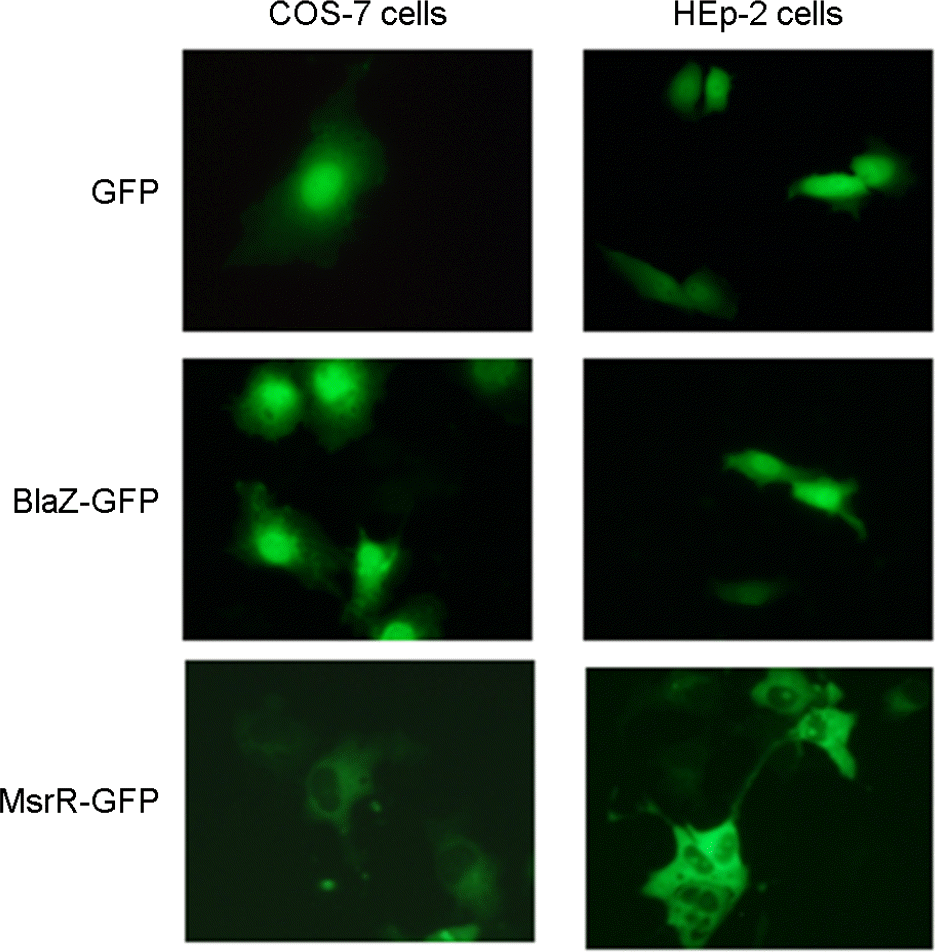 | Figure 1.
Nuclear targeting of fusion proteins of BlaZ-GFP in host cells. COS-7 and HEp-2 cells were transfected with blaZ and msrR cloned with pAcGFP1-N2 vector, respectively. Cells were transfected with the empty pAcGFP1-N2 vector as a control. Expression of BlaZ-GFP fusion proteins in host cells was observed using fluorescence microscope. Magnification: ×40. |
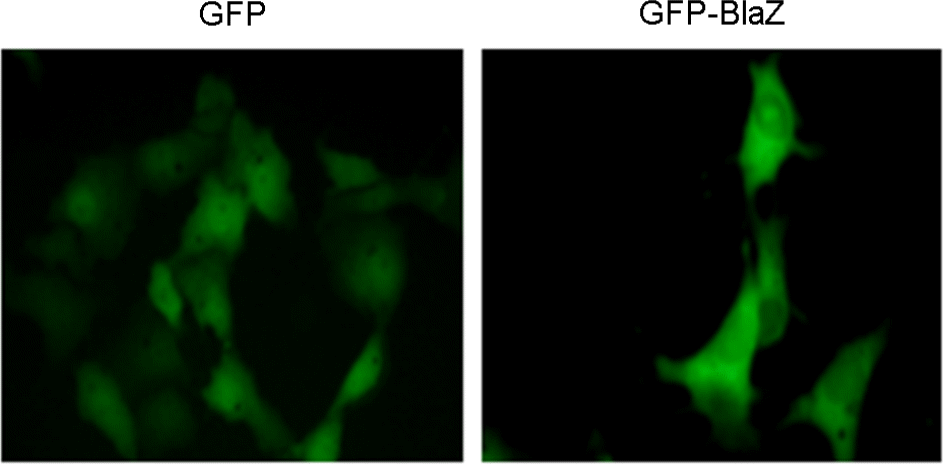 | Figure 2.
Cytosolic localization of fusion proteins of GFP-BlaZ in host cells. COS-7 cells were transfected with pAcGFP1-C2 vector or blaZ cloned with pAcGFP1-C2 vector. Expression of GFP-BlaZ fusion proteins in host cells was observed using fluorescence microscope. Magnification: ×40. |
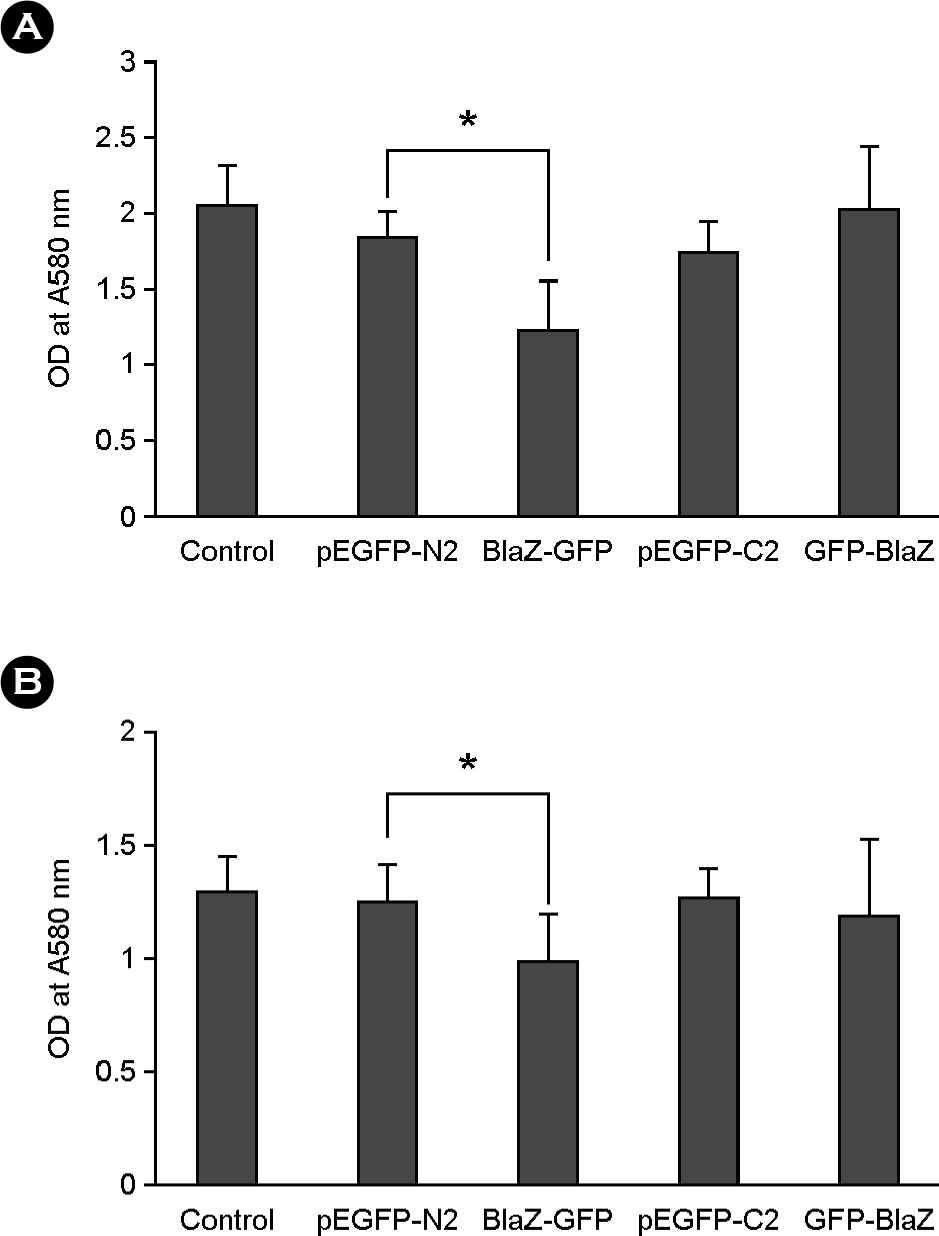 | Figure 3.
Cytotoxicity of β-lactamase fused with GFP in host cells. Cells were transfected with plasmid constructs of blaZ cloned with pAcGFP1-C2 and pAcGFP1-N2 vector, respectively. Cells were transfected with pAcGFP1-N2 or pAcGFP1-C2 vectors as a control. Cells transfected cells with plasmid constructs were incubated at 37°C for 44 h, and MTT assay was performed. (A), COS-7 cells. (B), HEp-2 cells. Data are presented as the mean ± SD of duplicate experiments. *p < 0.05. |
Table 1.
Prediction of NLS sequences in functional proteins of S. aureus
Table 2.
Prediction of NLS sequences in hypothetical proteins of S. aureus




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download