Abstract
Multilocus sequence typing (MLST) of Salmonella is useful method for replacing serotyping using antisera but is limited by difficulties associated with in polymerase chain reaction (PCR). We optimized the PCR reaction, especially annealing temperature and extension time (94°C for 2 min; 40 cycles at 94°C for 30 sec, 56.8°C for 1 min, 72°C for 2 min; and 72°C for 10 min). The degradation of PCR product by thermostable nucleases was inhibited by using template DNAs treated proteinase K or purified by a commercialized preparation kit. The resulting modified MLST was used as accurate and fast typing method.
Go to : 
REFERENCES
1). Grimont PAD, Weill FX. Antigenic formulae of the Salmonella serovars. 9th ed.Paris, France: WHO Collaborating Centre for Reference and Research on Salmonella, Institut Pasteur website;Available at:. http://www.pasteur.fr/ip/portal/action/WebdriveActionEvent/oid/01s-00036-089.
2). Fitzgerald C, Collins M, Van Duyne S, Mikoleit M, Brown T, Fields P. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J Clin Microbiol. 2007; 45:3323–34.
3). O'Farrell B, Haase JK, Velayudhan V, Murphy RA, Achtman M. Transforming microbial genotyping: A robotic pipeline for genotyping bacterial strains. PLoS One. 2012; 7:e48022.
4). Gibson JR, McKee RA. PCR products generated from unpurified Salmonella DNA are degraded by thermostable nuclease activity. Lett Appl Microbiol. 1993; 16:59–61.
5). Dahiya S, Kapil A, Kumar R, Das BK, Sood S, Chaudhry R, et al. Multiple locus sequence typing of Salmonella Typhi, isolated in north India-a preliminary study. Indian J Med Res. 2013; 137:957–62.
6). Seong WJ, Kwon HJ, Kim TE, Lee DY, Park MS, Kim JH. Molecular Serotyping of Salmonella enterica by complete rpoB gene sequencing. J Microbiol. 2012; 50:962–9.
7). Hyeon JY, Chon JW, Park JH, Kim MS, Oh YH, Choi IS, et al. A comparison of subtyping methods for differentiating Salmonella enteric serovar Enteritidis isolates obtained from food and human sources. Osong Public Health Res Perspect. 2013; 4:27–33.
8). Achtman M, Hale J, Murphy RA, Boyd EF, Porwollik S. Population structures in the SARA and SARB reference collections of Salmonella enterica according to MLST, MLEE and microarrary hybridization. Infect Genet Evol. 2013; 16:314–25.
9). Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012; 8:e1002776.
10). Gu W, Yang Z, Chen Y, Yin J, Yang J, Li C, et al. Molecular characteristics of Salmonella enterica Paratyphi A in Yunnan Province, southwest China. Infect Genet Evol. 2015; 30:181–5.
Go to : 
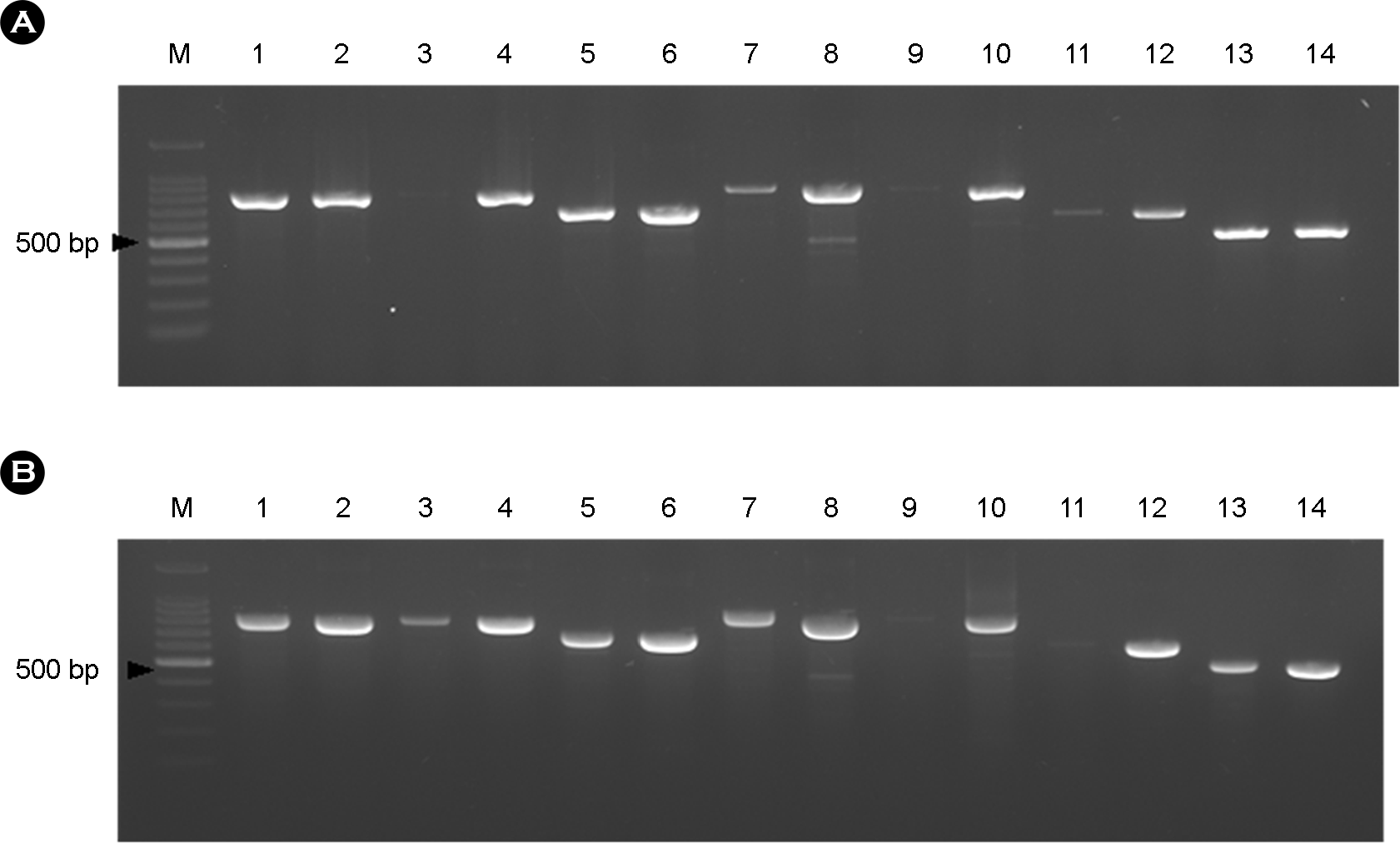 | Figure 1.
Comparison of the amplified PCR products using two commercialized PCR premix kits. Aliquots of 5 μl of PCR products were analyzed on a 1.5% agarose gel at 100 volts for 30 min. The PCR products with the optimized condition showed thicker and cleaner bands than those obtained by the previous condition with two different kits. (A) iNtRON Co., Ltd. PCR premix, Korea (B) SNC Co., Ltd PCR premix, Korea. M: 100 bp of DNA marker; Lanes 1~2, aroC, amplified with the previous and optimized conditions; Lanes 3~4: dnaN, amplified with the previous and optimized conditions; Lanes 5~6, hemD, amplified with the previous and optimized conditions; Lanes 7~8, hisD, amplified with the previous and optimized conditions; Lanes 9~10, thrA, amplified with the previous and optimized conditions; Lanes 11~12, sucA, amplified with the previous and optimized conditions; Lanes 13~14, purE, amplified with the previous and optimized conditions. |
Table 1.
Oligonucleotide sequences of primers for multilocus sequence type
Table 2.
Alleles numbers and sequence types of 20 strains of Salmonella serovars




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download