Abstract
Salmonella causes a wide variety of diseases ranging from mild diarrhea to severe systemic infections, such as like typhoid fever, in multiple organisms, ranging from mice to humans. A lack of ptsI, which encodes the first component of phosphoenolpyruvate (PEP): carbohydrate phosphotransferase system (PTS), is known to cause Salmonella Typhimurium attenuation; however, the mechanisms behind this have not yet been elucidated. In this study, a DNA microarray was performed to determine why the virulence of ptsI mutants is attenuated under low-oxygen conditions in which the ptsI expression is enhanced. Of 106 down-regulated genes, the most repressed were pdu and tdc genes, which are required for propanediol utilization and threonine and serine metabolism, respectively. In addition, half the flagellar genes were down-regulated in the ptsI mutant strain. Because pdu genes are induced during infection and Tdc products and flagella-mediated motility are necessary for the invasion of S. Typhimurium, the invasive ability of ptsI mutants was examined. We found that ptsI mutation reduced the ability of S. Typhimurium to invade into host cells, suggesting that reduced expression of the pdu, tdc, and flagellar genes is involved in the attenuation of ptsI mutants.
Go to : 
REFERENCES
1). Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007; 85:112–8.
2). Finlay BB, Brumell JH. Salmonella interactions with host cells: in vitro to in vivo. Philos Trans R Soc Lond B Biol Sci. 2000; 355:623–31.
3). Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001; 52:259–74.

4). Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol. 2008; 6:613–24.

5). Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010; 8:401–12.

6). Clements M, Eriksson S, Tezcan-Merdol D, Hinton JC, Rhen M. Virulence gene regulation in Salmonella enterica. Ann Med. 2001; 33:178–85.
7). Rhen M, Dorman CJ. Hierarchical gene regulators adapt Salmonella enterica to its host milieus. Int J Med Microbiol. 2005; 294:487–502.
8). Poncet S, Milohanic E, Mazé A, Nait Abdallah J, Aké F, Larribe M, et al. Correlations between carbon metabolism and virulence in bacteria. Contrib Microbiol. 2009; 16:88–102.

9). Le Bouguénec C, Schouler C. Sugar metabolism, an additional virulence factor in enterobacteria. Int J Med Microbiol. 2011; 301:1–6.

10). Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, et al. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008; 10:958–84.
11). Götz A, Eylert E, Eisenreich W, Goebel W. Carbon metabolism of enterobacterial human pathogens growing in epithelial colorectal adenocarcinoma (Caco-2) cells. PloS One. 2010; 5:e10586.

12). Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol Microbiol. 2003; 47:103–18.
13). Bowden SD, Hopper-Chidlaw AC, Rice CJ, Ramachandran VK, Kelly DJ, Thompson A. Nutritional and metabolic requirements for the infection of HeLa cells by Salmonella enterica serovar Typhimurium. PloS One. 2014; 9:e96266.
14). Bowden SD, Rowley G, Hinton JC, Thompson A. Glucose and glycolysis are required for the successful infection of macrophages and mice by Salmonella enterica serovar typhimurium. Infect Immun. 2009; 77:3117–26.
15). Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006; 70:939–1031.

16). Kok M, Bron G, Erni B, Mukhija S. Effect of enzyme I of the bacterial phosphoenolpyruvate: sugar phosphotransferase system (PTS) on virulence in a murine model. Microbiology. 2003; 149:2645–52.
17). Shin D, Cho N, Kim YJ, Seok YJ, Ryu S. Up-regulation of the cellular level of Escherichia coli PTS components by stabilizing reduced transcripts of the genes in response to the low oxygen level. Biochem Biophys Res Commun. 2008; 370:609–12.
18). Lim S, Yun J, Yoon H, Park C, Kim B, Jeon B, et al. Mlc regulation of Salmonella pathogenicity island I gene expression via hilE repression. Nucleic Acids Res. 2007; 35:1822–32.
19). Kim MJ, Lim S, Ryu S. Molecular analysis of the Salmonella typhimurium tdc operon regulation. J Microbiol Biotechnol. 2008; 18:1024–32.
20). Lonnstedt I ST. Replicated microarray data. Statistica Sinica. 2002; 12:31–46.
21). Plumbridge J. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr Opin Microbiol. 2002; 5:187–93.
22). Plumbridge J. A mutation which affects both the specificity of PtsG sugar transport and the regulation of ptsG expression by Mlc in Escherichia coli. Microbiology. 2000; 146:2655–63.
23). Zeppenfeld T, Larisch C, Lengeler JW, Jahreis K. Glucose transporter mutants of Escherichia coli K-12 with changes in substrate recognition of IICB (Glc) and induction behavior of the ptsG gene. J Bacteriol. 2000; 182:4443–52.
24). Yew WS, Gerlt JA. Utilization of L-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. J Bacteriol. 2002; 184:302–6.
25). Zhang Z, Aboulwafa M, Smith MH, Saier MH Jr. The ascorbate transporter of Escherichia coli. J Bacteriol. 2003; 185:2243–50.
26). Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol. 1997; 179:6633–9.
27). Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol. 1999; 181:5967–75.
28). Conner CP, Heithoff DM, Julio SM, Sinsheimer RL, Mahan MJ. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Nati Acad Sci U S A. 1998; 95:4641–5.
29). Adkins JN, Mottaz HM, Norbeck AD, Gustin JK, Rue J, Clauss TR, et al. Analysis of the Salmonella typhimurium proteome through environmental response toward infectious conditions. Mol Cell Proteomics. 2006; 5:1450–61.
30). Klumpp J, Fuchs TM. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology. 2007; 153:1207–20.
31). Kim M, Lim S, Kim D, Choy HE, Ryu S. A tdcA mutation reduces the invasive ability of Salmonella enterica serovar typhimurium. Mol Cells. 2009; 28:389–95.
32). Lim S, Kim M, Choi J, Ryu S. A mutation in tdcA attenuates the virulence of Salmonella enterica serovar Typhimurium. Mol Cells. 2010; 29:509–17.
33). Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS microbiol Rev. 2003; 27:505–23.

34). Landini P, Zehnder AJ. The global regulatory hns gene negatively affects adhesion to solid surfaces by anaerobically grown Escherichia coli by modulating expression of flagellar genes and lipopolysaccharide production. J Bacteriol. 2002; 184:1522–9.
35). Haiko J, Westerlund-Wikstrom B. The role of the bacterial flagellum in adhesion and virulence. Biology. 2013; 2:1242–67.

36). Eichelberg K, Galán JE. The flagellar sigma factor FliA (sigma(28)) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect Immun. 2000; 68:2735–43.
37). Olsen JE, Hoegh-Andersen KH, Casadesús J, Rosenkranzt J, Chadfield MS, Thomsen LE. The role of flagella and chemotaxis genes in host pathogen interaction of the host adapted Salmonella enterica serovar Dublin compared to the broad host range serovar S. Typhimurium. BMC microbiol. 2013; 13:67.
38). Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993; 57:543–94.
39). Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000; 64:694–708.
40). Hesslinger C, Fairhurst SA, Sawers G. Novel keto acid formate-lyase and propionate kinase enzymes are components of an anaerobic pathway in Escherichia coli that degrades L-threonine to propionate. Mol Microbiol. 1998; 27:477–92.
Go to : 
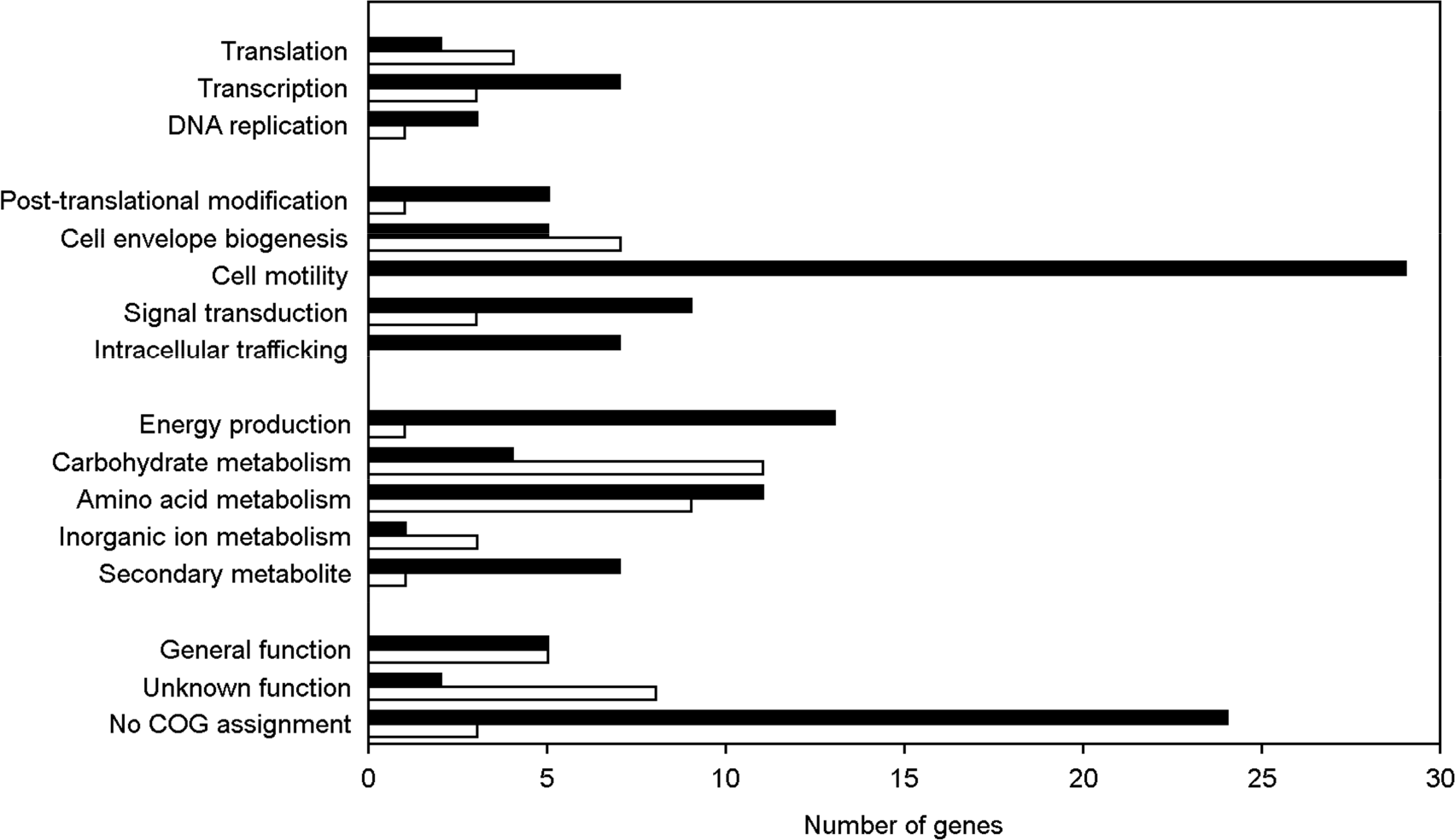 | Figure 1.Distribution of differentially expressed genes. The genes whose expression was changed in the ptsI mutant were grouped into COG functional categories. The bars (black, down-regulated; and white, up-regulated) represent the number of genes in each COG category. |
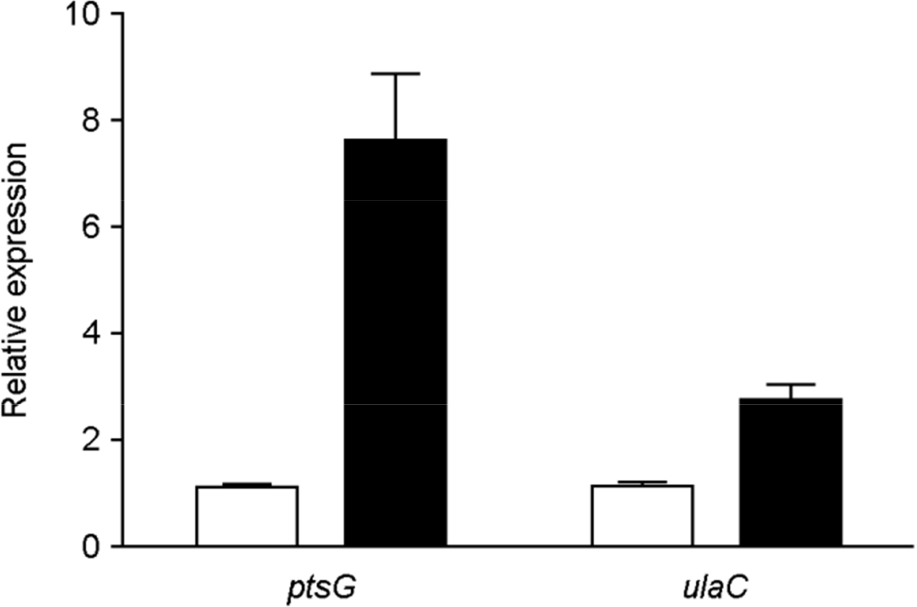 | Figure 2.mRNA levels of ulaC (STM2344) in mlc mutants. Bacterial cultures were grown statically to mid-log phase. After total RNA isolation, qRT-PCR analysis was performed to determine ptsG and ulaC transcript levels. mRNA levels of each gene in the wild type were arbitrarily set to 1 (white bars), and then the relative expression of each mlc mutant gene (black bars) was determined by normalizing the mRNA levels to this value. Error bars indicate the standard deviations obtained in three independent experiments performed in duplicate. |
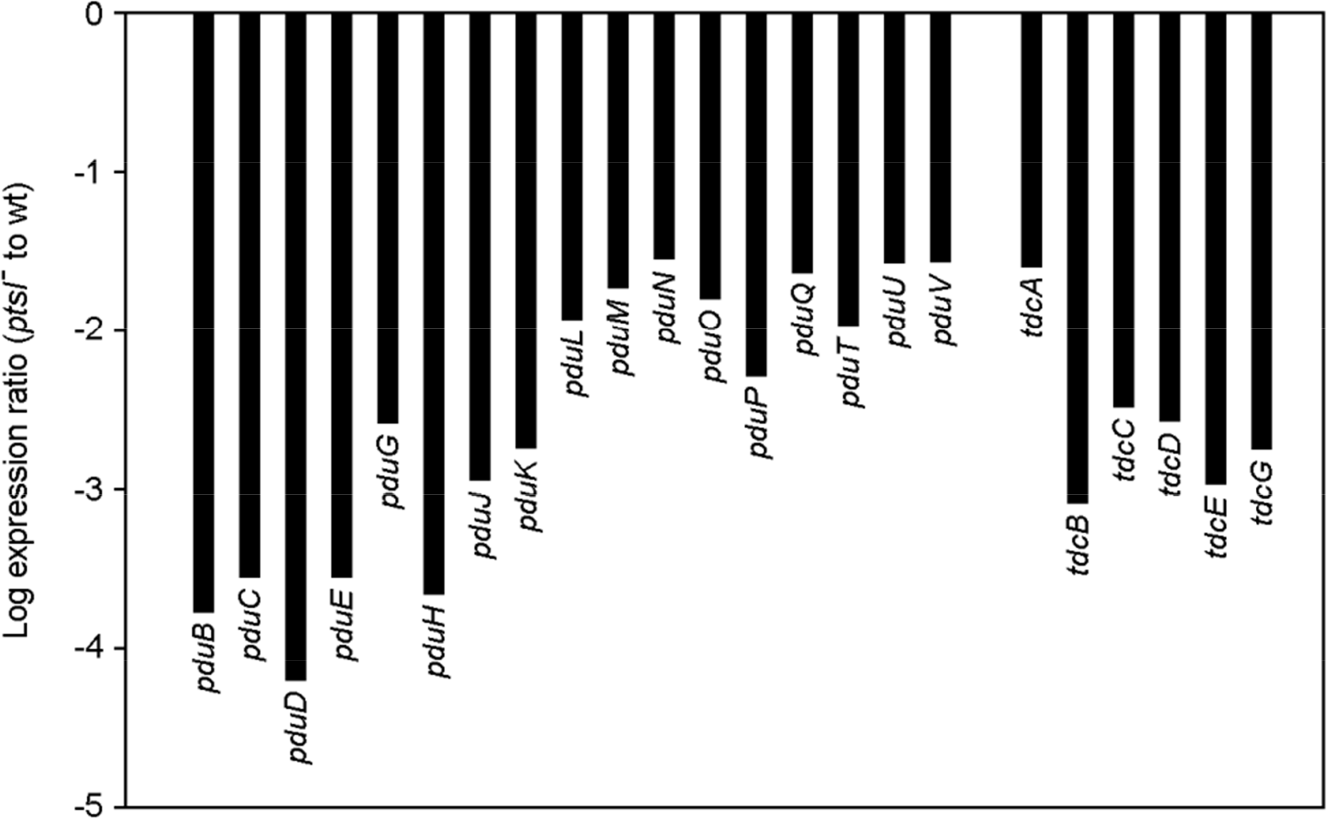 | Figure 3.The fold change for the pdu and tdc genes. Bars represent the expression ratio (ptsI− to wt) for each gene, which was derived from the log2-transformed microarray hybridization results. |
 | Figure 4.The fold change for the flagella and chemotaxis genes. Bars represent the expression ratio (ptsI− to wt) for each gene, which was derived from the log2-transformed microarray hybridization results. |
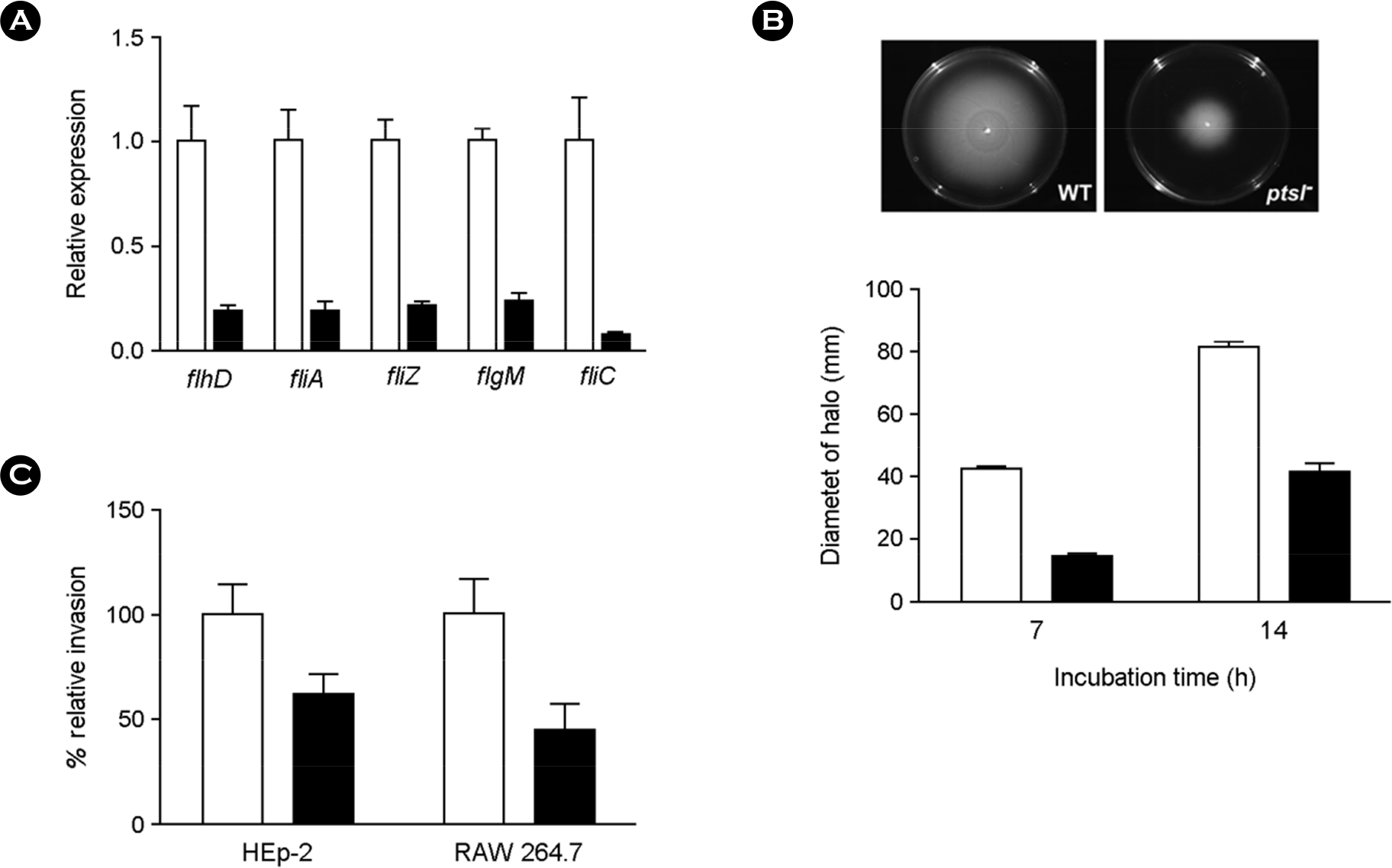 | Figure 5.Defects in flagella biosynthesis in ptsI mutants. In all bar graphs, white and black bars represent the wild-type (WT) and ptsI mutant (ptsI−) mutant strains, respectively. (A) Validation of microarray results for five flagellar genes. mRNA levels were measured through qRT-PCR, and the mRNA levels of each gene in WT were set to 1. Error bars indicate the standard deviations (SD) obtained in three independent experiments performed in duplicate. (B) WT and ptsI− were compared for motility. Bar graphs show the mean halo diameters (zones of motility) ± SDs obtained from a quantitative motility assay performed in triplicate. The zone of motility was measured at 7 and 14 h after inoculation. (C) Salmonella invasion into HEp-2 epithelial and RAW 264.7 macrophage cells. The mammalian cells infected by WT and ptsI− were lysed 2 h after infection and dilutions of the suspension were plated on LB agar medium to determine CFU. Data are presented as percentages of CFU of WT. Values are the means ± SDs obtained in three independent experiments performed in duplicate. |
Table 1.
List of primers used in qRT-PCR
Table 2.
The genes induced by ptsI mutation
Table 3.
List of the highly induced genes in ptsI mutants
Table 4.
List of the highly induced genes in ptsI mutants




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download