Abstract
Hepatitis C virus (HCV) is known to be a major cause of chronic hepatitis, liver cirrhosis and hepatocarcinoma. Therapeutic reagents are improving, but are still limited, and the protective vaccine against HCV is not available yet. However, the research of HCV life cycle and pathogenesis has been difficult due to obstacles, which are the lack of effective cell culture systems and small-animal models. Recently, breathtaking progress in terms of HCV replication system has been made using various forms of HCV clones and human hepatocarcinoma 7 cell lines (huh 7). The establishment of complete cell-culture system for HCV replication gave researchers opportunities to study the entire viral life cycle including entry, assembly, release of viral particles and the interaction with host cells. In fact, these efforts now appear to move into the identification and the development of innovative anti-HCV reagents. In this review, we go over the biological characters of HCV, a variety of in vitro cell culture, in vivo animal models of HCV infection, HCV immune-pathogenesis and the application of HCVcc system in terms of developing anti-HCV reagents.
Go to : 
REFERENCES
1). Seong MH, Kil H, Kim JY, Lee SS, Jang ES, Kim JW, et al. Clinical and epidemiological characteristics of Korean patients with hepatitis C virus genotype 6. Clin Mol Hepatol. 2013; 19:5–50.

2). Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000; 132:296–305.

3). Atug O, Akin H, Yilmaz Y, Sari M, Tozun N. Pegylated interferon/ribavirin-induced sudden sensorineural hearing loss in a patient with chronic hepatitis C. J Gastrointestin Liver Dis. 2009; 18:256.
4). Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002; 76:13001–14.

5). Mondada D, Pache I, Leopold K, Thorens J, Moradpour D, Gonvers JJ. Hepatology. Rev Med Suisse. 2006; 2:218–20. 223-6. 228–30.
6). Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wölk B, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007; 446:801–5.

7). Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, et al. Binding of hepatitis C virus to CD81. Science. 1998; 282:938–41.

8). Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002; 21:5017–25.

9). D'Souza ED, O'Sullivan E, Amphlett EM, Rowlands DJ, Sangar DV, Clarke BE. Analysis of NS3-mediated processing of the hepatitis C virus non-structural region in vitro. J Gen Virol. 1994; 75:3469–76.
10). Fournier C, Sureau C, Coste J, Ducos J, Pageaux G, Larrey D, et al. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J Gen Virol. 1998; 79:2367–74.
11). Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudoparticles containing functional E1-E2 envelope protein complexes. J Exp Med. 2003; 197:633–42.

12). Ikeda M, Yi M, Li K, Lemon SM. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol. 2002; 76:2997–3006.

13). Kato T, Date T, Miyamoto M, Furusaka A, Tokushige K, Mizokami M, et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology. 2003; 125:1808–17.

14). Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005; 11:791–6.

15). Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc Natl Acad Sci U S A. 2006; 103:7408–13.

16). Lindenbach BD, Meuleman P, Ploss A, Vanwolleghem T, Syder AJ, McKeating JA, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc Natl Acad Sci U S A. 2006; 103:3805–9.
17). Bettauer RH. Chimpanzees in hepatitis C virus research: 1998–2007. J Med Primatol. 2010; 39:9–23.

18). Xie ZC, Riezu-Boj JI, Lasarte JJ, Guillen J, Su JH, Civeira MP, et al. Transmission of hepatitis C virus infection to tree shrews. Virology. 1998; 244:513–20.

19). Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998; 4:1065–7.

20). Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, et al. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001; 7:927–33.

21). Li K, Li NL, Wei D, Pfeffer SR, Fan M, Pfeffer LM. Activation of chemokine and inflammatory cytokine response in hepatitis C virus-infected hepatocytes depends on Toll-like receptor 3 sensing of hepatitis C virus double-stranded RNA intermediates. Hepatology. 2012; 55:666–75.

22). Takahashi K, Asabe S, Wieland S, Garaigorta U, Gastaminza P, Isogawa M, et al. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc Natl Acad Sci U S A. 2010; 107:7431–6.

23). Arnaud N, Dabo S, Maillard P, Budkowska A, Kalliampakou KI, Mavromara P, et al. Hepatitis C virus controls interferon production through PKR activation. PLoS One. 2010; 5:e10575.

24). Cho H, Kang H. The interaction between HCV-infected huh7.5 cells and HCV-specific T cells. Korean J Microbiol. 2014; 50:169–72.
25). Hall CH, Kassel R, Tacke RS, Hahn YS. HCV+ hepatocytes induce human regulatory CD4+ T cells through the production of TGF-beta. PLoS One. 2010; 5:e12154.
26). Sulkowski MS, Kang M, Matining R, Wyles D, Johnson VA, Morse GD, et al. Safety and antiviral activity of the HCV entry inhibitor ITX5061 in treatment-naive HCV-infected adults: a randomized, double-blind, phase 1b study. J Infect Dis. 2014; 209:658–67.

27). Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med. 2011; 17:589–95.

28). Nettles JH, Stanton RA, Broyde J, Amblard F, Zhang H, Zhou L, et al. Asymmetric binding to NS5A by daclatasvir (BMS-790052) and analogs suggests two novel modes of HCV inhibition. J Med Chem. 2014; 57:10031–43.

Go to : 
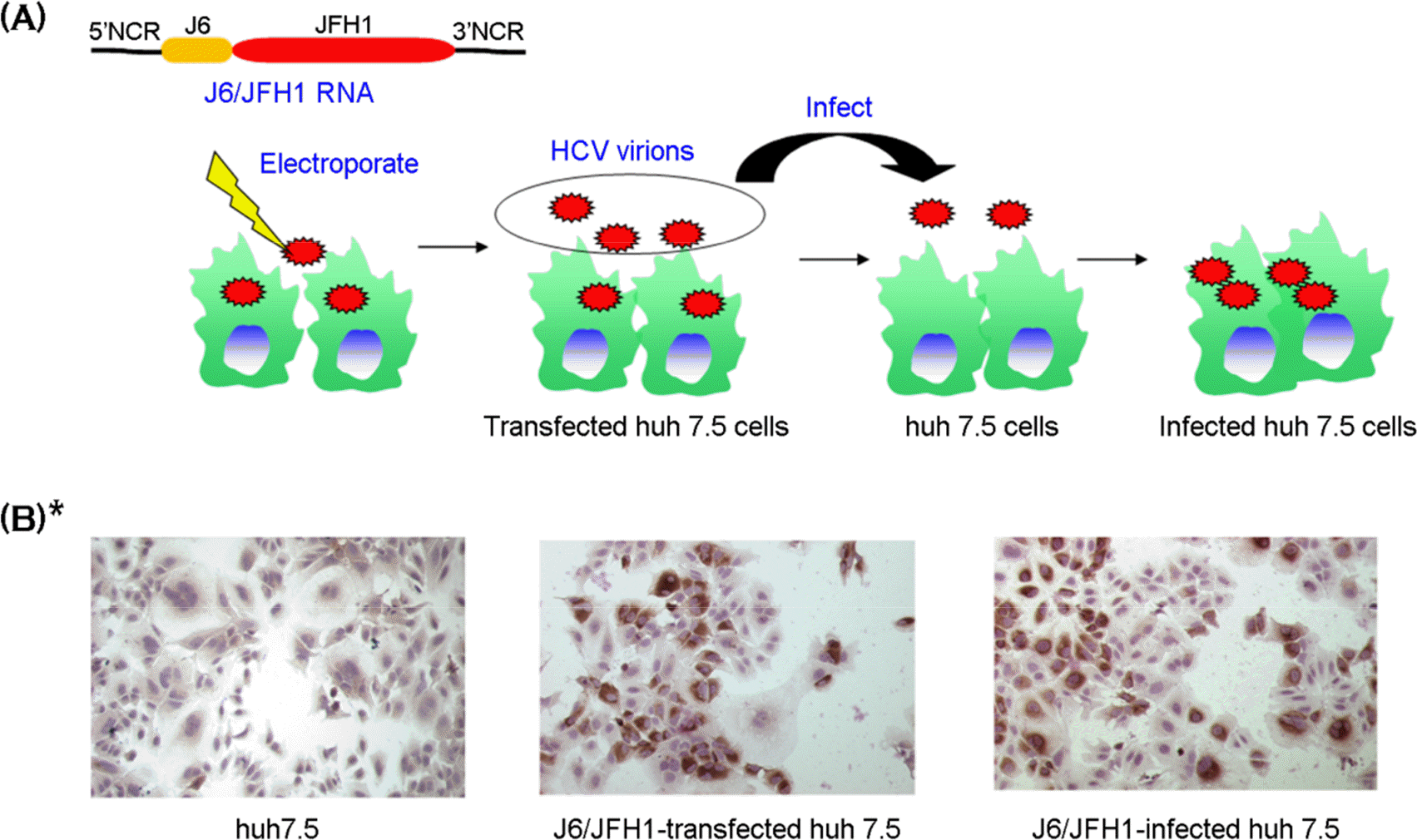 | Figure 1.
Establishment of infectious HCV cell culture system with J6/JFH1 (HCVcc). (A) The experimental outline of HCVcc. (B) Immunohistochemical expression of HCV NS5A protein (dark spots) in cytoplasm of huh 7.5 cells. ∗(B) is modified from Cho et al. (24). |
Table 1.
In vitro and in vivo models to investigate HCV pathogenesis




 PDF
PDF ePub
ePub Citation
Citation Print
Print


 XML Download
XML Download